Patterning the butterfly wing through Wnt signaling
Posted by Tirtha Das Banerjee, on 31 August 2023
A recent paper in Science Advances titled ‘Spatial and temporal regulation of Wnt signaling pathway members in the development of butterfly wing patterns’ explores the expression and function of Wnt signaling pathway members in setting up butterfly wing patterns. We caught up with first author Tirtha Das Banerjee and corresponding author Antόnia Monteiro from the National University of Singapore to learn about the behind the paper story.

What was known about Wnt signalling and the butterfly wing patterning before your work?
Tirtha: Wnt signalling is a fundamental signalling pathway that regulates cell communication, cell growth, and cell proliferation in metazoans. A lot is known about this pathway in classical model systems such as Drosophila, but little is known in other systems, such as in butterfly wings. In butterfly systems a few Wnt ligands, primarily WntA and Wnt1, had been associated with the development of bands and eyespots, but most of the other ligands in the pathway, and their receptors, had not been examined in any species.
Antónia: Back in 2006, in my lab at Buffalo, we had visualized Wg/Wnt1 (using an antibody against the Wnt1 protein in humans) at the center of butterfly eyespots, but it wasn’t until we were able to produce a transgenic line, expressing two copies of wg back to back, that folded upon each other when transcribed, that we were able to knock-down this gene to observe its effects on eyespots. A graduate student in my lab in Singapore, Nesibe Özsu, worked on this, and she was able to see smaller eyespots developing when the double stranded RNA was transcribed inside cells via a heat-shock. Tirtha, however, used CRISPR to try and get stronger phenotypes on the wing.
How did this project get started? And Tirtha, what brought you to Antónia’s lab?
Tirtha: This work was partially inspired by my previous work on venation patterning published in Development in 2020 where I observed a very dynamic pattern of Armadillo (Arm) in the larval wings of butterflies. Since Arm is an important component of Wnt signaling, I hypothesized that the ligands (the Wnts) and their receptors (the Frizzleds) might also show dynamic patterns of expression as the wing develops. I started examining their expression, one by one, and uncovered that these other components of Wnt signaling are also extremely dynamic across both larval and pupal wings.
I visited Antónia’s lab back in 2014 during a summer internship program from my graduate studies at NIT-Durgapur. Back then, I visualized the expression of two transcription factors, Engrailed and Spalt, using antibodies that produced some interesting patterns. Even though these early stains were extremely blurry and saturated with colors, they were super cool to me. I proposed an hypothesis for butterfly venation patterning based on the data but due to the limited time of my internship (of 2 months), I was unable to continue the work. Later after graduation I continued this work, which later got published. I was hooked into the evo-devo of wing vein (and color) patterning ever since.
Why did you choose the butterfly wing as a model to undercover the complexities of Wnt signalling?
Tirtha: Developing butterfly wings are simple 2D sheets of cells where numerous ligands, receptors, signal-transducers, and transcription factors orchestrate the specification of extremely complex patterns (of colour) that will be visible in the adult wing. Cells send and receive cues, and interact with each other during development, to specify these colour patterns. Since larval and pupal wings are miniature versions of adult wings, it is easy to map the molecules involved in these signalling processes to the final colour patterns they are likely affecting.
Antónia: I started working on butterfly wing patterns as an honours student in 1990. I thought butterflies would make great genetic and developmental models because they can produce a ton of eggs and the wings are large and flat, like Tirtha said. In addition, they were much prettier than Drosophila, and their intricate colour patterns were the real hook.
Can you summarise your key findings?
Tirtha: In our recent work we found that the expression and function of different Wnt signaling members varies quite a bit during wing development. For example, Frizzled4, one of the Wnt receptors, is expressed quite uniformly during larval development, but is missing from the future eyespot centers, where canonical Wnt signaling, mediated by Armadillo, is taking place. During the pupal stage, however, the expression of Frizzled4 completely changes and now it is co-expressed in the eyespot centers together with Armadillo. During these two stages Frizzled4 is also likely playing different roles: it is involved in the localization of the eyespot centers during the larval stage because its removal leads to eyespot center duplications, and it is involved in wing scale orientation during the pupal stage. We observed similar dynamics for many other genes we tested in the study and proposed mechanisms of how these genes are likely interacting to specify the multiple cell fates on the wings of butterflies.
Antónia: A key realization for me was observing that a patchwork of different Frizzled ligands and receptors is expressed across the whole wing, which means that every cell of the wing is either producing or receiving Wnt signals, but processing them in different ways. How this patchwork of different Wnt ligands and receptors work together will be interesting to investigate in future.
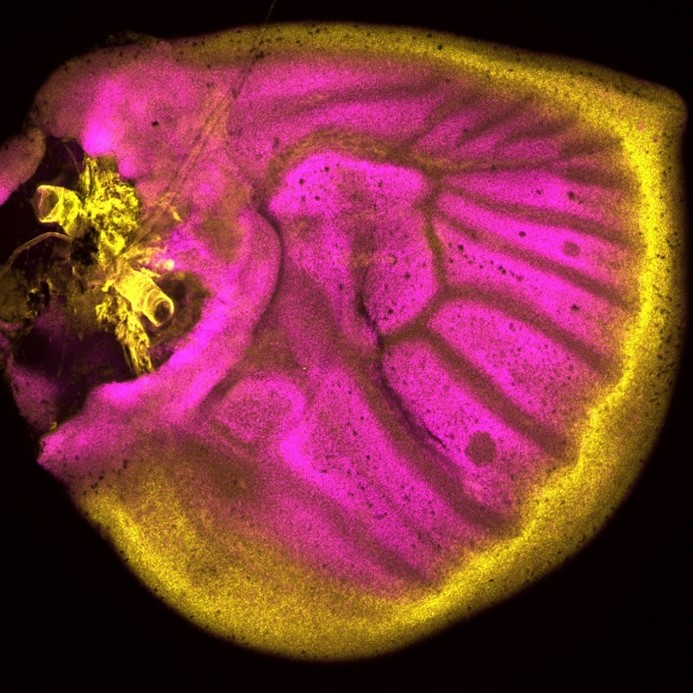
Did you have any particular result or eureka moment that has stuck with you?
Tirtha: Well one of the exciting moments was when I observed the armadillo CRISPR phenotype showing a double eyespot on the wing. I was also very excited the day I observed the different frizzled patterns on the pupal wings. It made me wonder how nature, over the course of evolution, has been intricately patterning wings by activating certain genes at some location while repressing them in other locations. It’s really incredible, and makes you sit down and think how things which we consider simple are so complex and elegantly tuned at the molecular level.
And the flipside: were there any moments of frustration or despair?
Tirtha: Ohh there were many. As a scientist I believe we all have accepted that there will be more moments of frustration than excitement. For example, for one of the Wnt ligands called Wnt1, I tried to knock it out with over ten different CRISPR guides, and injected over 5000 embryos. I got no results. Doing stainings with the traditional enzymatic in-situ hybridization was also a painstaking job. I am glad at least those days are over with the new HCR technique we have adapted in the lab.
A recent Development paper also used CRISPR to look at WntA and Frizzled receptors in the butterfly wing patterning. How does the two papers complement each other?
Tirtha: The study from Arnaud Martin’s lab is extremely impressive. Their lab has consistently produced papers that have advanced the field of biological colour pattern evolution. The authors have generated a massive amount of information in different species of butterflies on how WntA is likely being transduced via the Frizzled2 receptors. They have also gathered functional data on other receptors such as frizzled, frizzled3, and frizzled4 patterning different aspects of the adult wing scales and venation which are not present in our work. The use of RNAi as a gene knockdown methodology they used would also be extremely useful for other labs working on similar lepidopteran tissues. The gene expression data from their lab and our lab confirms the presence of the different receptors during larval and pupal stages in similar conserved domains strengthening our hypothesis that these receptors are involved in patterning adult wings across butterflies.
Where will this story take the lab?
Antónia: Well, we have only touched the tip of iceberg with the present study. This work has opened up many new avenues for new students to work on. For example, a student in the lab is now testing a hypothesis we proposed on the interaction of Frizzled4 and Arm in larval wings and the function of frizzled2 and frizzled9 in the development of scales and colour patterns. Another student is testing where the rest of the Wnt signalling members are expressed and what function they play. A postdoc in the lab is trying to visualize whether there is a Wnt1 morphogen gradient around the eyespot centers, as hypothesized nearly 45 years ago.
Tirtha, what’s next for you?
Tirtha: Well, I am currently involved in the development of more advanced technologies for spatial transcriptomics that will allow us to multiplex the number of genes we tested in these studies. Basically, we would be able to understand how perturbations to individual members of Wnt signaling affect the expression of other Wnt pathway members or downstream targets. I hope the upcoming work will have broad applicability across different model systems.


 (No Ratings Yet)
(No Ratings Yet)