The people behind the papers – Clément Dubois, Shivam Gupta, Andrew Mugler and Marie-Anne Félix
Posted by the Node Interviews, on 12 July 2021
This interview, the 95th in our series, was published in Development earlier this year.
Cell migration needs to be precisely regulated during development so that cells stop in the right position. A new paper in Development investigates the robustness of neuroblast migration in the C. elegans larva in the face of both genetic and environmental variation. To hear more about the story, we met the paper’s four authors: Clément Dubois and Shivam Gupta, and their respective supervisors Andrew Mugler (currently Assistant Professor at the Department of Physics and Astronomy at the University of Pittsburgh, where his lab recently moved from Purdue University) and Marie-Anne Félix (Principal Investigator at Institut de Biologie de l’Ecole Normale Supérieure in Paris and Research Director at CNRS).

Andrew and Marie-Anne, can you give us your scientific biographies and the questions your labs are trying to answer?
AM: After a PhD and postdoctoral research on theoretical biological physics, I began my own position investigating noise in biological systems. Cells contend with unavoidable noise from environmental fluctuations, small molecule numbers and many other sources. My group uses tools from theoretical physics to uncover strategies that cells use to precisely sense their environment, undergo controlled developmental or phenotypic changes, and execute collective behaviours.
M-AF: After a PhD in cell biology, I wanted to combine evolutionary biology with cell/developmental biology. In Paul Sternberg’s lab at Caltech and in my lab’s early years, we compared vulva development in different nematode species, giving flesh to the concept of developmental systems drift. Over the past 20 years, we moved to a microevolutionary scale: studying the sensitivities of developmental systems to various types of perturbation and comparing these with their evolutionary variation. The overall thread is to assess how much development biases phenotypic evolution, with some phenotypes being more easily reached than others upon random genetic variation or quantitative tuning of parameters.
Studying natural variation led us to seek C. elegans in nature, which was a lot of fun. With many colleagues, we have since been probing its population genetics structure, and the zoo of its associated microbes. Bringing real worms back to the lab also led us to discover natural variation in the duration of their multigenerational memory.
Clément and Shivam – how did you come to work in your respective labs and what drives your research today?
CD: I did an MSc in ecology and evolution, and was specifically interested in host-pathogen interactions. After a first internship in the Pasteur Institute working on mosquito-virus interactions, my former supervisor suggested I apply to Marie-Anne’s lab. I started working on the intraspecific evolution of C. elegans resistance to microsporidia, and then did a year as a technician working on the evolution of a peculiar cell in C. elegans called P3.p. This year was really formative. I gained a lot of scientific maturity, and realised how rich and friendly the C. elegans community is. Then Marie-Anne proposed I do a PhD on the evolution of QR neuroblast migration. Considering the different topics I worked on, I would say that my research is mostly driven by curiosity and the question of how much genotypes can shape phenotypes.
SG: I came to Purdue University as a grad student in 2015. I joined Dr Mugler’s lab after doing a lab rotation to get a feel for the research. The research work in the Mugler lab was very challenging, and it was intriguing to see how fundamental physics can explain the behaviours of microscopic organisms.
How has your research been affected by the COVID-19 pandemic?
M-AF: The lab closed for 3 months in the spring of 2020; since then it has been somewhat restricted, but we can do experiments. Of course, C. elegans is a great organism to work on because we do not lose strains: they can be kept frozen or can live for 3 months in diapause (a pre-adaptation to SARS-CoV2!). But some projects were affected for more than 3 months. To enhance social interactions, since the fall we decided to meet after lunch every day in a hybrid format (real life and video). This compensates for the usual lunch-time exchanges, be they scientific or not. On the positive side, the pandemic also gave us the impetus to meet more readily with colleagues worldwide, attending each other’s lab meetings or organising informal meetings.
AM: As a theory group, we could continue most of our work from home without interruption. However, developing ideas in any area benefits from true collective brainstorming, which has been slowed without the ability to physically meet at the whiteboard. Additionally, the pandemic has increased my childcare responsibilities, which has left less time to dedicate to students. On the positive side, I moved institutions in the middle of the pandemic, and I now hold hybrid group meetings with group members from both places, which is perfectly natural to do virtually in these times.
How did your labs come to collaborate on this project?
AM & M-AF: This was all started by our colleague Rik Korswagen, who has been working on C. elegans QR migration as a model. He contacted us both to write an application for a collaborative grant from the Human Frontier Science Program. We got the grant, and this has been a great experience. The modelling of body size variation was not in the grant, but the collaboration took place naturally once we were networking.
Can you give us the key results of the paper in a paragraph?
CD, SG, AM & M-AF: The goal was to measure the precision and natural variation in the final position of a cell that migrates during development. Other studies in various systems have examined the directionality of cell migration, but to our knowledge not the precision in the cell’s final position. Specifically, we studied two descendants of the QR neuroblast lineage that migrate a long way during C. elegans development. The Korswagen lab had shown that the end of their migration is specified by a temporal rather than a spatial mechanism. Given this temporal regulation, we hypothesised that a change in body size would affect the final position of the neurons relative to body landmarks. Indeed, we observed that a smaller body size resulted in the cell migrating further. Cell position even changed as a function of maternal age, a factor previously known to affect body size. We developed a mathematical model of the expectation, taking into account larval growth during the migration. The data did not fit quantitatively the simplest form of the model. Then, a model with partial compensation of body size, grounded on measurements of cell speed, fitted the experimental results without any free parameters. Finally, we revealed natural variation among wild isolates of C. elegans in the neurons’ final position, large enough that they result in a change in their neighbouring cells.
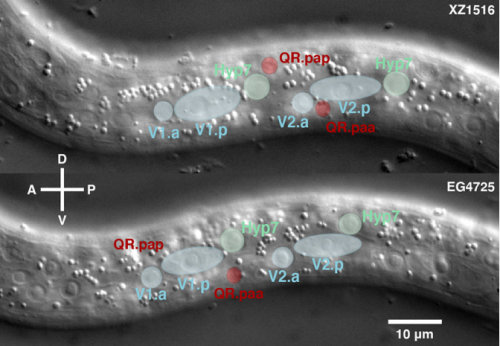
Do you have any idea why cell migration speed is dependent on body size?
CD & M-AF: The first that comes to mind is that a larger body size means larger cells (at least for a species such as C. elegans, which has a quasi-invariant number of cells). But interestingly, we could not find much published on whether a larger cell would go faster or slower than a smaller one. Both appear possible, depending on specific motility mechanisms. Another possibility is that the external environment of the cell is altered as a function of body size; for example, a change in the Wnt gradient that influences QR migration, either its initial concentration or its decay length.
On the other hand, in the wild isolates, the eventual position of QR.pax did not correlate with body size: why do you think this is?
CD & M-AF: We did not detect a significant correlation, and we cannot rule there would be one with a larger set of experiments. In any case, this means that much of the variation is due to other factors that can go in another direction than the effect of body size. Variation in many genes and processes can affect a quantitative trait such as cell position.
Your paper was published in Development through the Review Commons route – how did you find the experience?
CD, SG, AM & M-AF: Great! This was our first time. With Review Commons, the manuscript is sent to reviewers without any submission to a specific journal. The reviewers do not have any specific journal in mind. In our case, the reviews were very constructive, especially by suggesting that we assessed the effect of maternal age. Once the reviews arrived, the authors can contact one of the participating journals, specifying what is planned for responding to reviews. Thanks to Development, this went very well.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
CD: The first eureka moment was to see the preliminary results on the natural variation of QR.pax final position. To be able to detect differences between wild isolates for the first time was really exciting and promising. From then on, I was excited at the end of each experiment after making plots from the data (and I spent a lot of time on them!).
SG: The first few months of my PhD work was mainly building mathematical models for temporal precision in gene expression. At first, I was a bit pessimistic that the rules of physics would explain the complex behaviour of organisms. After analysing experimental data from C. elegans, it was very satisfying to see that the behaviour of microscopic cells can be explained by our models.
And what about the flipside: any moments of frustration or despair?
CD: I did not have a particularly memorable moment of frustration or despair – even negative results were informative. Nonetheless, the most challenging part was probably the measurements of embryo and larva lengths at different time points, in parallel on four genotypes. It required a lot of organisation and efficacy, but it was worth it.
SG: To solve complex biological problems, we start with a simple model and slowly build up the model. After a point, our model became very complex and it took days to run the computer program. It was frustrating to wait days to see results, but in the end, when results were promising, it was very joyful.
What next for you two after this paper?
CD: In the short term I’m doing two things: writing my PhD manuscript and working on the intraspecific evolution of the final position of QR.pax. Using Recombinant Inbred Lines between CB4932 and JU1242 (two genotypes with an opposite phenotype), I found a QTL on chromosome IV. The goal now is to find the polymorphism associated with the difference in QR.pax final position.
SG: I have successfully defended my thesis and secured a job in the pharmaceutical industry. My ambition is to advance drug discovery and safety of drugs through developing pharmacokinetics/pharmacodynamics models and analysing clinical trial data.
Simple mathematical models can prove very useful for sharpening basic understanding of biological systems
Where will this story take the Mugler and Félix labs?
AM: This has been one of the most positive demonstrations that simple mathematical models can prove very useful for sharpening basic understanding of biological systems; I hope to continue in this vein with this and other collaborations.
M-AF: I hope we can find the polymorphism behind the divergent phenotypes of two wild C. elegans isolates. Beyond, I will keep vulva development as our main developmental model, but, as both are regulated by the same Wnt gradient, we will likely come back again to QR migration.
Finally, let’s move outside the lab – what do you like to do in your spare time in Paris and West Lafayette?
CD: Paris is full of small and hidden restaurants that are really good. I like to explore them with my partner. I also enjoy spending time with my friends playing board games, climbing in the gym or in Fontainebleau, a forest close to Paris known for its large boulders.
SG: I enjoy exploring new places around West Lafayette for hiking and biking.
AM: Biking, exploring new restaurants and travelling.
M-AF: Hunting. For nematodes.


 (No Ratings Yet)
(No Ratings Yet)