The people behind the papers – Rémi-Xavier Coux & Ruth Lehmann
Posted by the Node Interviews, on 5 April 2018
Development and homeostasis depend crucially on the maintenance of cell identity, and in gamete-producing tissues the somatic/germline distinction is paramount. A recent paper in Development explores how cell identity is secured in the Drosophila ovary by studying the function of the conserved tumour suppressor L(3)mbt. To find out more about the story, we caught up with first author Rémi-Xavier Coux and his supervisor Ruth Lehmann of the Skirball Institute at New York University School of Medicine.

Ruth, can you give us your scientific biography and the questions your lab is trying to answer?
RL I grew up in Germany and received my PhD in the lab of Christiane Nuesslein-Volhard. I was in her lab at the very beginning when the genes required for establishing embryonic polarity and pattern were identified. My project involved the genetic analysis of gap genes and a group of maternal effect genes, we termed the posterior group of genes, as mutation in these genes affect the development of the embryonic abdomen and my cytoplasmic transplantation experiments suggested a gradient of patterning activity emanating from the posterior pole. I became intrigued by this posterior pole plasm as it is also the site of germ cell formation in Drosophila. So, I made the more unusual and often discouraged decision to stay with my graduate project, yet decided to delve into molecular analysis of these genes. By the time I arrived for my first independent position at the Whitehead Institute at MIT in 1988, I had molecular entry points for the identification of the nanos, oskar and pumilio genes in my suitcase.
Analysis of these genes was really exciting as it showed that the mRNAs encoding Oskar and Nanos were localized to the posterior pole and that the translation of these RNAs was specifically regulated such that only the localized RNA (and now we know that is only a very small fraction of the total RNA) is translated while unlocalized RNA is translationally repressed. From the beginning, I have been amazingly lucky to have incredibly talented graduate students and postdocs in my lab, not only did they make all these discoveries but they also taught me a lot.
In 1996, I was recruited to the newly founded Skirball Institute at NYU School of Medicine. Here my lab focused completely on germ cell biology studying the germ line life cycle. We are particularly interested in three areas:
1. How the germ line-soma dichotomy is initially established in the early embryo. Critical for the initial distinction between soma and germ line are the properties of membraneless germ granules that co-ordinate the posttranscriptional regulation of RNAs specifically needed for germ cell formation, specification, transcriptional silencing and germ cell migration to the somatic gonad. Crucial for progress here has been the development of ever so powerful imaging modalities (from light sheet microscopy and super resolution microscopy to electron microscopy) and the development of many different ways to mark molecules and observe them in vivo.
2. How interactions between cells from different origins coordinate growth and differentiation of the gonad so that primordial germ cells mature into germ line stem cells and deposit eggs during adult life. Initially, we relied on forward genetic screens to address this big and fascinating problem of organ development but now we are increasingly take advantage of whole genome genetic analysis by RNAi and Crispr/Cas9 as well as high-throughput molecular analysis such as single cell sequencing at different developmental stages.
3. Finally, we are interested in how the unique role of germ cells as the only cells of the body with the potential to give rise to a complete new organism manifests specialized adaptions. So, we have become intrigued by the broadest sense of mutual ‘host-pathogen’ interactions as they relate to the germ line. Here, we are interested in how germ line regulatory mechanisms manage to control transposable elements activity, the interplay between genome and mitochondria during the germ line life cycle and emerging optional relationships like the ability of the intracellular bacterium Wolbachia to grow in the Drosophila host without direct harm. We reason, that such germline specific control and defence mechanisms protect the germ line, but may also provide opportunity for species to evolve via changes to the germline.
And Rémi-Xavier, how did you come to be involved with this project?
RXC l(3)mbt was identified in the laboratory of Elisabeth Gateff, who was the first to use Drosophila to identify tumor-suppressor genes (Gateff, 1978, Gateff et al., 1993). As the name says, ‘l(3) malignant brain tumor’ mutations cause brain tumors in Drosophila larvae. Many years later, Chris Yohn, a postdoc in our lab, identified several alleles of l(3)mbt in a clonal screen for maternally expressed genes that affected germ cell formation in the embryo. While Chris determined that the PGC formation defect was a secondary consequence of L(3)mbt’s role in early embryonic nuclear divisions, he also observed that l(3)mbt females were viable at the permissive temperature (brain tumors form only at high temperature) but they were completely sterile and produced no eggs. This suggested an additional role for L(3)mbt in gonadogenesis (Yohn et al., 2003). The project rested for a while, until Cayetano Gonzalez and his group at the IRB in Barcelona published an intriguing observation. The Gonzalez lab profiled l(3)mbt larval brain tumors and showed that in these tumors many germline genes were apparently derepressed (Janic et al., 2010). We were intrigued by this possible soma-to-germline transformation, so when I joined the lab as a graduate student I started two projects on l(3)mbt. First, I asked whether l(3)mbt brain tumor cells indeed behaved like bona fide germ cells and could be used to identify novel regulators of the germline fate. Second, I wanted to test whether the sterility phenotype was also due to a soma-to-germline transformation. Our paper describes the results of the later study that proved much more successful than the former.
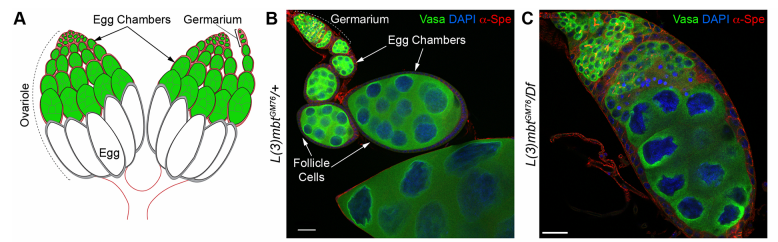
Can you give us the key results of the paper in a paragraph?
RL&RXC In addition to its role in suppressing brain tumors in the larvae, L(3)mbt functions in two tissues of the ovary to safeguard oogenesis: the somatic support cells and the germline. We found that in both tissues, L(3)mbt prevents expression of genes incompatible with normal development: in the somatic ovarian cells, L(3)mbt represses genes normally expressed in the germline while in the germ cells, it silences testis and neuronal genes. This, and the fact that l(3)mbt mutant tissues still express genes characteristic of the tissue of origin revealed a function broader than previously thought. Our study therefore suggests that L(3)mbt functions as a tissue specific transcription repressor rather than simply silencing germline genes in somatic tissues. L(3)mbt binding sites overlap with insulator elements (Richter et al., 2012) so it is also possible that L(3)mbt functions in insulator complexes.
Do you think L(3)mbt work with different partners in different tissues? Any ideas how it plays tissue-specific roles?
RL&RXC Indeed, L(3)mbt has been found to function with the dREaM/MMB and LINT complexes in S2 and Kc167 somatic embryonic cells (Georlette et al., 2007; Meier et al., 2012). However, our genetic studies suggest that L(3)mbt functions independently of dREaM in the ovary. Several components of the dREaM complex are required for endo-replication (polyploidization) of both somatic ovarian cells and nurse cells in the germline, but these processes seem not to require L(3)mbt function. It would be interesting to test if L(3)mbt functions in only one of these two chromatin complexes in other tissues besides the ovary and how the potential switch between complexes is regulated.
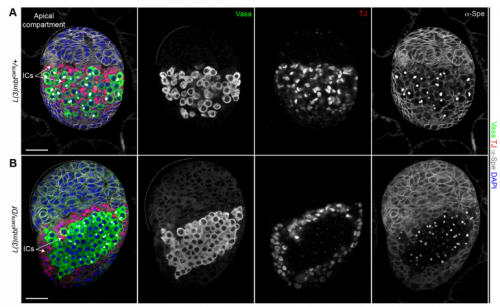
Your results suggest that rather than guarding against transdifferentiation, L(3)mbt guards against adoption of mixed identity. Where does this leave the concept/importance of transdifferentiation in development?
RL&RXC This is a conceptual question we really struggled with: the “orthodox” definition of transdifferentiation is complete acquisition of another cell identity. However, functionally testing fate switching in vivo is very challenging. Jarrault and colleagues beautifully showed that in C. elegans the Y epithelial cell transdifferentiates into a fully functional neuron (PDA) in wild-type larvae (Jarrault et al., 2008). It is one of the only examples of complete transdifferentiation to our knowledge. There are many more examples, especially with mutations in chromatin factors, where cell-specific gene signatures are misexpressed. At the end, it comes down to the assay that is used to define a ‘cell fate switch’. For example, can we call a cell that aberrantly expresses most of another cell type’s transcriptome trans-differentiated without a functional assay?
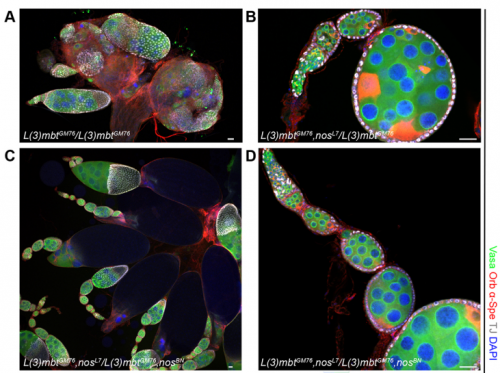
When doing the research, did you have any particular result or eureka moment that has stuck with you?
RXC Yes, I was very glad to observe that nanos mutations suppress the l(3)mbt mutant ovarian phenotypes. It was late at the confocal and I immediately emailed Ruth. The morning after, we checked that embryos laid by l(3)mbt, nos double mutant females had the typical patterning defects caused by nos mutations. When this control was done, we were really excited!!!
And what about the flipside: any moments of frustration or despair?
RXC I tried transplanting l(3)mbt tumorous brain cells into embryos devoid of germline to test if they could, at least partially, behave as germ cells. This experiment was quite challenging, and we could not detect the cells a few hours after transplantation, which was very frustrating! We now know that this experiment likely did not work because these neural-origin cells may not be completely transformed.
What next for you Rémi-Xavier?
RXC I just started a postdoc in the Cohen-Tannoudji and Navarro-Gil labs in the Stem Cell Biology and Development Department, Pasteur Institute in Paris. I will study and characterize Transcription Factor bookmarking in the early mouse embryo.
Where will this work take the Lehmann lab?
RL Further study of L(3)mbt may provide us with clues about the transcriptional mechanisms underlying germline-soma dichotomy. We found in our study that mutating a key regulator of germline fate, the translational repressor Nanos, suppresses the somatic gonadal defects of l(3)mbt mutant ovaries almost completely. Thus, Nanos targets may be key regulators that distinguish between the germline and soma program and the l(3)mbt mutants may guide us towards their identification.
Finally, let’s move outside the lab – what do you like to do in your spare time?
RXC In my free time, I enjoy music, modern art, playing rugby and outdoor activities such as sailing and mountain activities.
RL I like hiking with my Aussie Luke.
Rémi-Xavier Coux, Felipe Karam Teixeira, Ruth Lehmann. 2018. L(3)mbt and the LINT complex safeguard cellular identity in the Drosophila ovary. Development 2018 145: dev160721
This is #38 in our interview series. Browse the archive here


 (4 votes)
(4 votes)