The people behind the papers – Ximena Anleu Gil & Dominique Bergmann
Posted by the Node Interviews, on 18 July 2018
Asymmetric division is a widespread mechanism for generating cellular diversity during developmental patterning. The stomata of flowering plants are epidermal valves that regulate gas exchange, and provide an accessible system to investigate the mechanisms of asymmetric cell division both within and across species. A paper in the new issue of Development reports an investigation of the molecular control of this process during development in the forage grass Brachypodium distachyon. We caught up with co-first author Ximena Anleu Gil and her supervisor Dominique Bergmann, Professor of Biology at Stanford University, to hear more about the story.

Dominique, can you give us your scientific biography and the questions your lab is trying to answer?
DB I’ve been fascinated by development, and especially the relationship between asymmetric cell division and cell fate, for more than two decades. As a PhD student, this meant studying body axis formation in C. elegans. I still love the stunning microscopy possible with worm embryos, but I began to be interested in less invariant development—we know most organisms have some degree of flexibility (and uncertainty) in their developmental trajectories– and so, as postdoc, I moved to plants to ask about the relationships between cell polarity, asymmetric divisions, and cell identity in organisms with a great deal of plasticity, resilience, and environmental responsiveness.
The challenge, of course, in choosing to work on something with flexible development is that to have any hope of dissecting genetic networks or organizing principles you need to focus on some relatively simple decisions. For my lab, that has been the development of stomata (pores that mediate uptake of CO2 from the atmosphere in exchange for water vapor from the plant) in the epidermis of leaves. Most of the lab works on stomata in Arabidopsis, where stomata and their non-stomatal neighbors are the product of a simple epidermal lineage: we are fascinated by the stem cell-like asymmetric divisions at the beginning of this lineage that can be modulated to create leaves of different sizes with different numbers of stomata. These days, we are looking at the lineage through transcriptomic and epigenomic approaches, by developing imaging tools to monitor asymmetric divisions and also by collaborating with ecophysiologists to look at how development affects behaviors in the “real world” and vice versa.
Ximena, how did you come to join the Bergmann lab, and what drives your research?
MXAG I joined the Bergmann lab as a lab technician after I finished my college degree. I had been studying the heat shock response mechanism in plants and was looking for technician jobs in other plant labs to expand my lab skills and also experience real-life science before starting a graduate degree. I was extremely lucky because Dominique was looking for a technician at that time.
I define and re-define my motivations as I learn more about the incredibly complex and beautiful world of biology, but what never changes is my amazement with the natural world. Visually and intellectually, nature is a fascinating puzzle to try to decipher. And as I learn more about plants and their flexible but robust development, I have become deeply intrigued with trying to understand developmental decisions from a plant’s perspective. The plant kingdom is so diverse and important for our planet and society, I feel that we should spend more time thinking about it.
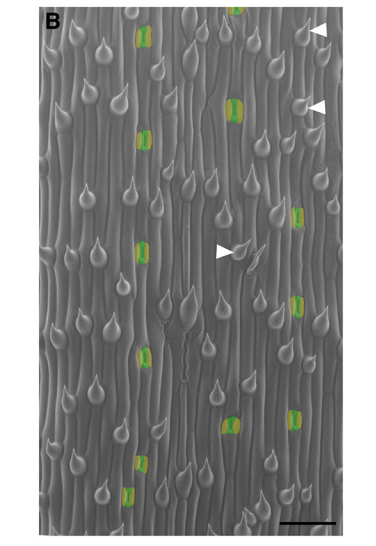
What makes Brachypodium a good (comparative) model for asymmetric cell division in development, and what were the gaps in your knowledge about the process before your current work?
DB & MXAG The stomatal lineage of Brachypodium, like those of rice, maize, wheat, and other grasses, is very organized. You can think of its development as a time-line where spatial position (base vs. tip of leaf) is a proxy for time. It’s like the Arabidopsis root in this way, which lends itself to certain simplifying behaviors relative to the randomly oriented self-renewing divisions of Arabidopsis. When YODA, the kinase at the heart of our recent Development paper, was first characterized, it was in the context of asymmetric divisions in the Arabidopsis embryo (by Wolfgang Lukowitz and colleagues) and the stomatal lineage. From these early studies, it was thought that the processes of physically asymmetric division and establishment of different cellular fates were intrinsically linked, and both controlled by YODA. Neither the Arabidopsis embryo (tiny and inaccessible for long-term imaging) nor the stomatal lineage (with its rounds of asymmetric division) could give us much clarity on the real connection between a physically asymmetric division and its immediate fate outcome. The beautifully ordered epidermis of Brachypodium allowed us to study them separately, both conceptually and experimentally. The presence of epidermal cell types not present in Arabidopsis like hairs and silica cells also gave us new things to measure, and it was exciting (but unexpected) that their patterning would also controlled by YODA. Finally, not inconsequentially, as challenging as it was to analyze the dwarfed bdyda1 mutants, atyda would have been worse because the plant is even tinier!
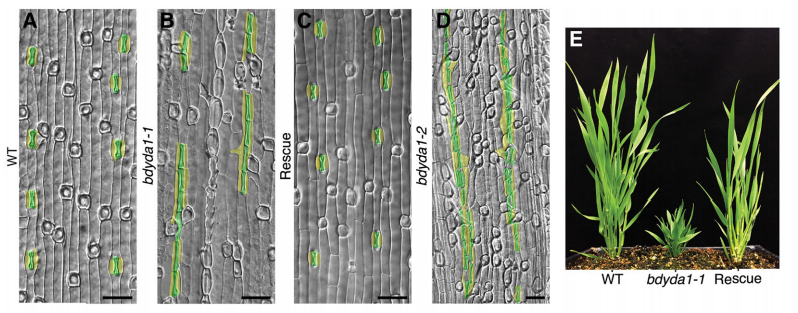
Can you give us the key results of the paper in a paragraph?
DB & MXAG Asymmetric cell divisions are feature of plants, animals and microbes and, in multicellular organisms, are critical to establish pattern and cell-type diversity. Much of our thinking about asymmetric divisions comes from flies and worms, where ideas about creating physical asymmetries and orienting the division of a mother cell has direct consequences for fate (through segregation of fate determinants or positioning of daughters near instructive cues). In our paper, we report the identification and characterization of a mutant isolated from a forward genetic screen based on its production of clustered stomata—typically a readout for defects in cell fate and asymmetric division. Through complementation experiments and CRISPR/Cas9 genome editing, we confirmed that the phenotypes were caused by mutations in the BdYDA1 gene. BdYDA1 is a very close homolog of AtYDA MAPKKK gene, a crucial member of a MAPK signalling cascade that is required to inhibit stomatal cell fate and enforce a patterning rule that prevents stomata from being direct neighbors in Arabidopsis. In contrast to loss of the Arabidopsis counterpart, bdyda1 mutants also displayed the same patterning defects in other non-stomata epidermal cells, including hair cells and silica cells. Surprisingly, each of these clustered patterns did not seem to arise from faulty physical asymmetric cell divisions but from improper enforcement of alternative cellular fates in these tissues. Our phenotypic analysis suggests that in Brachypodium, BdYDA1 works as part of a general molecular switch that controls cell identity to ensure proper spacing of epidermal cell types. They also helped expand our understanding of the mechanisms through which YODA genes are acting in the establishment of differential cell fates during plant development.
In terms of its upstream and downstream factors/signalling cassettes, how similar do you think BdYDA1 works compared to AtYDA?
DB & MXAG We have no reason to think that kinase activity differs between AtYDA and BdYDA1, however, each of these proteins has a large, less well conserved, N-terminal domain. In Arabidopsis, this domain regulates protein activity and is suspected to link AtYDA to upstream kinases, one of which has unique features in the Brassicas (mustard plant family, including Arabidopsis), so that suggests potentially different upstream inputs. On the other hand, homologues of the EPF family of signaling peptides that act upstream of AtYDA are found in grasses and overexpression of one of these in barley affects stomata (Hughes et al., 2017 doi.org/10.1104/pp.16.01844) so there is some potential for conservation, too.
Downstream, work in Arabidopsis revealed a core set of dedicated stomatal transcription factors that promote the entry into, and exit out of stem-cell like divisions, as well as guide the differentiation process of stomatal guard cells. The expression and protein stability of these transcription factors are highly regulated, and about a decade ago Ph.D. student Cora MacAlister and postdoc Greg Lampard found that the stomatal initiating factor SPEECHLESS (so named for the absence of stomata or “mouths”) was phosphorylated by MAPKs downstream of YODA. SPCH has been duplicated in Brachypodium and neither paralog encodes a protein with all the known MAPK sites present in AtSPCH, so it is possible that the BdSPCHs aren’t the ultimate target of BdYDA1. And we certainly have no idea what the targets in silica and hair cells might be! What is also really interesting is that homologues of the Arabidopsis polarity protein BASL, which is both target of MAPK phosphorylation and potential scaffold enabling differential AtYDA inheritance after asymmetric division, are not found at all in grasses. What, if anything, is used in BASL’s place?
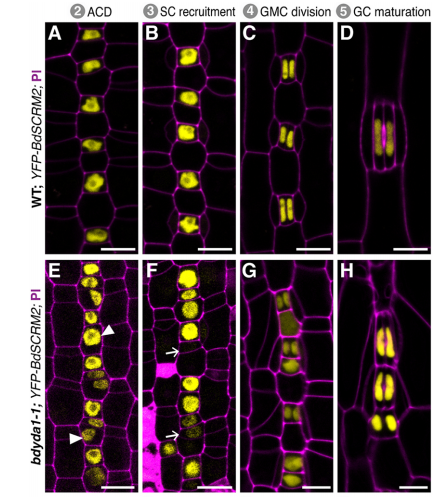
What does this work tell you more broadly about how cell fates are determined in plants?
DB & MXAG Unlike animal development, plant development mostly occurs post-embryonically and is heavily influenced by the environment. The Arabidopsis stomatal lineage for example, has the flexibility to modify the number of precursor cells in response to their environment, and it does so by regulating the number and types of asymmetric divisions. Environment can also mean “neighbor cells” and more and more evidence is accumulating from root development and grass stomatal subsidiary cell recruitment (Raissig et al., 2017, doi:10.1126/science.aal3254) that neighboring cells have a stronger role in providing the appropriate signals that determine cell fate. There are more questions than answers now, but our BdYDA1 work sheds some light into some of these inquiries. We propose a shift in our understanding of MAPK signaling during asymmetric cell division in plants, from being all about the physical asymmetry of cells before and after a division event to thinking about this process as a more dynamic one, i.e. one that requires enduring cross-talk between neighboring cells to ensure the correct fate of daughter cells after an asymmetric division event. Our thinking is that plant cells are locked in place and there is no programmed death and removal of cells; thus, what a cell becomes is of great significance to neighbors stuck beside it for life—it’s in the interest of these neighbors to provide cues to coax that cell into becoming something appropriate, and for that cell to take its time evaluating the incoming information.
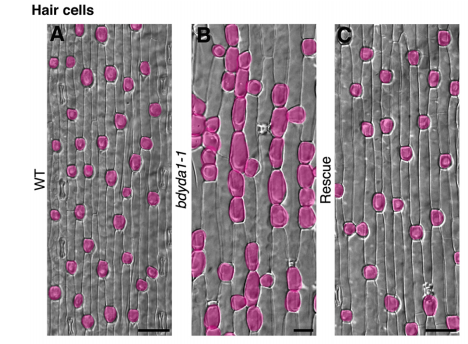
When doing the research, did you have any particular result or eureka moment that has stuck with you?
MXAG A BdYDA1 allele was identified in a screen that Emily Abrash and Juliana Matos (with Akhila Bettadapur) initiated back in 2011, and BdYDA1 was actually the first Brachypodium gene Emily cloned, using a “clone by guessing” strategy—the plant phenotypes of excessive stomata and compact growth were so reminiscent of AtYDA mutants that a homologue seemed like an obvious choice. But after Emily confirmed that she had a mutation in BdYDA1, the project was put on the back burner for a while because the phenotypes were so superficially similar in atyda and bdyda1 mutants that we doubted we’d learn anything new. It wasn’t until we created the cell identity markers (BdSPCH2-YFP, BdMUTE-YFP and BdSCRM2-YFP) and tools for live cell imaging that it became clear that something was different! I was tasked with finishing this project started by the original Brachy team, and at the beginning was a little nervous about re-analyzing Emily’s data and re-evaluating her interpretations. I spent a lot of time in the microscope looking at the epidermis of WT and bdyda1 plants and developed a feeling that asymmetric cell divisions looked just fine in bdyda1. But, since this observation ran counter to the dogma in the field, initially I didn’t feel so confident about my observations, nor did I fully understand the implications this would have for our understanding of stomatal development. After many, many meetings with Dominique and other lab members, presenting in lab meeting, attending conferences, and finally putting this project in paper format, I gained confidence. Then things just came full circle! I wouldn’t say it was a eureka moment because I had to convince myself and others that what I was observing was real and reproducible.
And what about the flipside: any moments of frustration or despair?
MXAG Of course! My self-confidence was shaken quite a bit during this time… worries that I was being too slow doing experiments, reading relevant papers, writing the paper, etc. I had done research as an undergrad and had had my own projects, but nothing of this magnitude. I had to learn many, many things like taking breaks, balancing my time working independently with my role as technician, and re-motivating myself when academic science felt too far away from the real world, among other things. I couldn’t have done it without such a supportive PI and lab. I was extremely lucky to have friends who reminded me about the importance of taking care of myself and having fun while doing research! More concretely, working with bdyda1 is quite difficult. The plants are so small and they don’t always want to be photographed… so trying and trying until I got the picture I needed and wanted without a doubt tested my patience and diligence.
What next for you after this paper?
MXAG I’m extremely excited to start my PhD! I’ll be joining the Plant Biology group at UC Davis where I hope to continue exploring plant development.
And where will this work take the Bergmann lab?
DB The initial idea for moving to Brachypodium was to use genetic screens to probe novelties in the grass stomatal lineage, like the rigid, file-like organization of stomata and their precursors and the presence of subsidiary cells flanking the stomatal guard cells. These screens, however, told us that the massive changes in stomatal form and pattern were due to minor rewiring of the core genetic components (e.g., Raissig et al., 2016 and 2017). This changed my attitude toward the experimental approach and the value of Brachypodium. We are unlikely to do more forward genetic screens, but rather will use Brachypodium (and CRISPR/Cas) to test ideas about conserved or divergent developmental mechanisms. I’m intrigued by a couple of projects we have going that suggest subfunctionalized, or even opposite, roles of Arabidopsis and Brachypodium genes in stomatal development. These studies might give us insight into protein evolution, or might reveal how the different developmental strategies of Arabidopsis and Brachypodium place different demands on the same protein activity. But I’m really excited that while the Bergmann lab may be streamlining its Brachypodium projects, former postdoc Michael Raissig, will be expanding the Brachypodium universe in his own new group at the University of Heidelberg in Germany.
Finally, let’s move outside the lab – what do you like to do in your spare time in California?
DB I love being outside in California, in all its seasons and diverse places: the mountains, the ocean, the forests. It’s magnificent to be out biking and hiking, and also eating and drinking products of this state’s agricultural bounty.
MXAG: I also enjoy spending time outside, particularly going to the ocean. I love the Pacific Northwest coastline because is so peaceful and mesmerizing. I’m also a big foodie! So, cooking, going to restaurants and farmer’s markets, eating… yeah I do a lot of that here.
Conservation and divergence of YODA MAPKKK function in regulation of grass epidermal patterning
Emily Abrash, M. Ximena Anleu Gil, Juliana L. Matos, Dominique C. Bergmann
Development 2018 145: dev165860 doi: 10.1242/dev.165860
This is #46 in our interview series. Browse the archive here.


 (1 votes)
(1 votes)