BSDB Gurdon Summer Studentship Report – Rufus Morgan
Posted by Rufus Morgan, on 5 January 2022
Regionalising the nervous system: the steps to produce spinal cord.
The nervous system is one of the first structures that develops during embryogenesis. However, the process by which regional identity is established in this system remains unclear and is a long-standing unanswered question of developmental biologists. The Metzis lab aims to elucidate some of the molecular mechanisms defining regional identity in the nervous system during its initial development. To address these questions, the group uses embryonic stem cells (ESCs) as an in vitro model of neural development combined with high throughput sequencing approaches and comparisons to in vivo mouse models. Previous research has demonstrated that the generation of spinal cord requires the activity of the transcription factor CDX2 (1, 2), but it is still unknown exactly how this happens at the molecular level. My project focussed on identifying potential downstream targets of CDX2, using a combination of experimental and computational approaches.
The Hox family are a set of transcription factors that have fascinated developmental biologists for decades due to their role in patterning the body plan and their conservation across Bilateria (animals with left-right symmetry). Interestingly, it’s found that these genes have distinct rostro-caudal domains of expression in the developing spinal cord (3) and are expressed in a colinear fashion (4). Hox genes are regulated by CDX transcription factors (5, 6), so the main aim of my project was to predict which Hox genes are directly regulated by CDX2during the emergence of spinal cord. This could elucidate potential Hox genes that act in conjunction or as a result of Cdx2 upregulation to promote spinal cord identity.
I initially approached this project by processing and analysing high throughput sequencing data from previous mRNA-seq experiments (7). This allowed me to identify gene expression patterns that occur during the differentiation of pluripotent ESCs into hindbrain versus spinal cord progenitors in vitro. I then evaluated these predictions by performing RT-qPCR to confirm that the published data from the mRNA-seq dataset matched the in vitro differentiations being conducted in the lab using the same protocol. I found that in general the predictions made were reproducible (Figure 1), so I finally went about verifying if CDX2 occupied the Hox genes that I had predicted, by analysing ChIP-seq datasets from previous studies (2, 8).
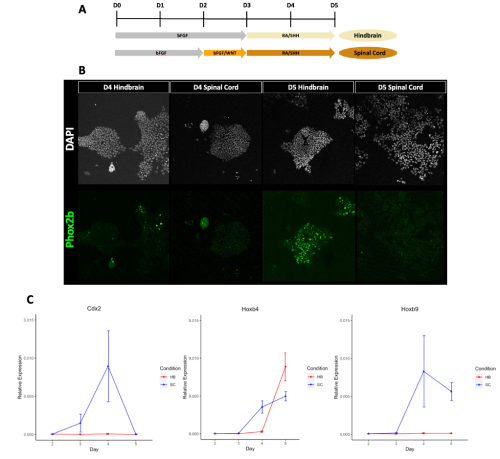
It was exciting but challenging completing my own independent research project for the first time, and it forced me to step outside my comfort zone and grow quickly. A good example of this was using 384-well plates for the first time during qPCR. It was daunting to plan each plate, calculate the reagent needed and pipette each one out in a reproducible manner. Initially it took me a long time and I made mistakes; nevertheless, I soon got the hang of it and could do them with my eyes closed! Furthermore, having had no previous experience in coding, I initially struggled to successfully output any results. However, the dry lab aspect of this project taught me to be perseverant and explore your options, as there is often more than one way to get to the correct result. Over this summer I have also learnt to think more deeply about why I am doing a specific experiment, what it can teach me, and what my next steps will be after obtaining the results.
Although difficult, I found my project in the Metzis lab thoroughly enjoyable, and the experience gained was invaluable. I learnt to always approach intimidating tasks quickly and get stuck in, which was also really helped by the fact that the lab team were supportive and would guide me through new techniques. I learnt to always have a plan B – it is unlikely everything will go perfectly the first time, so preparing for that eventuality is necessary to not slow down your workflow too much.
This project has opened my eyes to what a professional research lab is like but also the questions and challenges present in the field of developmental biology. It has encouraged me to think more deeply about what we don’t already know about this field, and potentially some unanswered questions that I could look to answer in the future. I have had an amazing summer at the Metzis lab, and I very much look forward to getting back to the bench to explore those unanswered questions!

1. Metzis V, Steinhauser S, Pakanavicius E, Gouti M, Stamataki D, Ivanovitch K, et al. Nervous System Regionalization Entails Axial Allocation before Neural Differentiation. Cell. 2018;175(4):1105-18.e17.
2. Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, et al. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci. 2013;16(9):1191-8.
3. Carpenter EM. Hox genes and spinal cord development. Dev Neurosci. 2002;24(1):24-34.
4. Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236(9):2454-63.
5. Amin S, Neijts R, Simmini S, van Rooijen C, Tan SC, Kester L, et al. Cdx and T Brachyury Co-activate Growth Signaling in the Embryonic Axial Progenitor Niche. Cell Rep. 2016;17(12):3165-77.
6. Neijts R, Amin S, van Rooijen C, Deschamps J. Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev Biol. 2017;422(2):146-54.
7. Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, et al. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12(8):e1001937.
8. Mahony S, Edwards MD, Mazzoni EO, Sherwood RI, Kakumanu A, Morrison CA, et al. An integrated model of multiple-condition ChIP-Seq data reveals predeterminants of Cdx2 binding. PLoS Comput Biol. 2014;10(3):e1003501.


 (2 votes)
(2 votes)