The people behind the papers – Qiang Zhu, Marçal Gallemí and Eva Benková
Posted by the Node Interviews, on 10 October 2019
This interview, the 69th in our series, was recently published in Development.
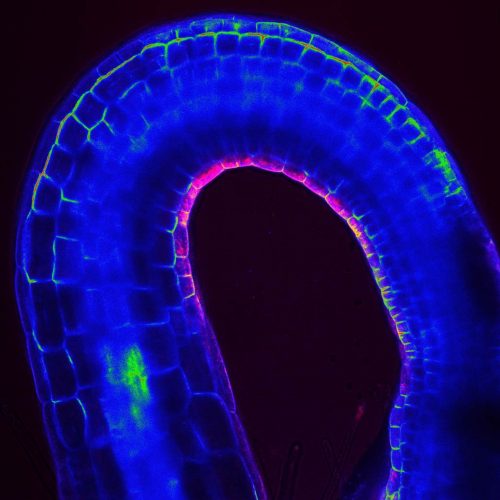
The apical hook is a transient structure that functions to protect the vulnerable apical meristem from damage when the seedling penetrates the soil. Although some of the molecular players regulating its development have been identified, many aspects have remained opaque, including how an early auxin asymmetry in the hypocotyl is established. A paper in Development now provides a link between hormone signalling and the gravitropic response of the seedling’s growing root in apical hook development. We caught up with co-first authors Qiang Zhu and Marçal Gallemí and their supervisor Eva Benková, Professor at the Institute of Science and Technology Austria in Klosterneuberg, to find out more about the project.

Eva, can you give us your scientific biography and the questions your lab is trying to answer?
EB I studied Molecular Biology and Genetics at the Masaryk University in the Czech Republic, and then obtained my PhD at the Institute of Biophysics of the Czech Academy of Sciences. As a postdoctoral fellow, I joined the laboratories of Professor Klaus Palme at the Max Planck Institute, Cologne, and Professor Gerd Jurgens at the University of Tübingen. During my PhD studies and later as a postdoctoral fellow I became interested in plant hormones and their exceptional impact on all aspects of plant development. I was fascinated by the simple and (among plant biologists) well-known experiment by Murashige and Skoog in reported 1957: they demonstrated that a modulation of the ratio between two plant hormones, auxin and cytokinin, can re-define the developmental programme of meristematic cells, resulting in either shoot or root formation. Thus, in 2007, when I got the opportunity to lead my independent research as group leader at the Plant Systems Biology department of the VIB in Belgium, I decided to pursue this topic and fully focus on plant hormones and the mechanisms underlying their cross-talk. In 2012, I moved to the Institute of Science and Technology Austria, where I continue this research line. My team is trying to dissect the molecular mechanisms underlying complex hormonal interactions and identify key points at which hormonal signalling pathways converge to control plant growth and development.
Qiang and Marçal, how did you come to work in the lab, and what drives your research today?
QZ As a plant biologist, I am impressed by how plants have developed complicated mechanisms that enable rapid and flexible adaptation of their growth and development to their ever-changing surrounding environment, and I’m curious about the mechanisms behind this. When I was in Ghent University, Eva and my previous lab had some joint projects on development of the apical hook, a very important transient structure for dicots when they grow out of the dark. At that time, I got to know this interesting model, and wanted to know more about it. It’s this interest in the topic and project that mainly drives my research. I am so lucky that I had the chance to join the Benková lab and work on the project. My research is greatly supported by Eva, who is talented, hard-thinking and dedicated. It’s really nice to work with her: we have a lot of discussions about this project, which also drive me to deeper investigation into the secret behind the formation of the apical hook.
MG Working in a lab, and specifically a molecular biology lab, is like playing games every day! We assemble and disassemble pieces of a puzzle trying to get to know what are they for, how they work and how they interact. Curiosity is what mainly drives my research but, with the current status of our planet, including extremely exploited natural resources, pollution and global warming, understanding how plants grow and adapt to the environment is also a key question for humanity.
Working in a lab…is like playing games every day
What was known about apical hook development before your current work, and in particular the role of gravity in the process?
EB The apical hook is developmentally a very important structure, which is transiently formed during germination to protect a delicate apical meristem and cotyledons from damage when seedlings penetrate through the soil. Several studies have reported that apical hook formation is driven by asymmetric distribution of auxin accumulating at the inner side of the hook. However, how this asymmetric pattern of auxin distribution is established, and which mechanisms determine formation of the apical hook during early phases of seedling germination, was largely unknown. Although previously there have been some indications that gravity might play a role in the apical hook development, for example from plants observed in space and from our own research, no systematic studies were reported. Compared with other plant developmental processes, research on the apical hook has lagged behind, probably due to the lack of a suitable technique that would enable observation of this process occurring in darkness. Luckily, with establishment of the dark-imaging system we could start to monitor the whole process of apical hook development in real time. I think this has been the most important technical advance and enabled us to address many interesting questions.
Can you give us the key results of the paper in a paragraph?
EB In our work, we showed that during early phases of germination, gravity-stimulated bending of the root acts as the initial cue to coordinate the formation of the apical hook. The core machinery mediating the root’s response to gravity is required for apical hook formation, and PIN2, which so far was considered to be a root-specific auxin efflux carrier, acts as an essential integrator of root-to-hypocotyl communication. We found that during early phases of germination, PIN2 activity is not restricted to the root: it manages transport of auxin to the hypocotyl, thus providing initial asymmetry in auxin distribution, which is further reinforced by the polar auxin transport machinery that is gradually established in the hypocotyl. Hence, two distinct developmental events taking place at opposite poles (root and shoot) of the plant axis are regulated by common regulatory machinery. Finally, we demonstrated that establishment of such inter-organelle transport system is a result of tight interplay between two hormonal pathways, abscisic acid (ABA) and gibberellins (GA), acting antagonistically during embryo maturation and early germination.
How do you think gravity-stimulated, auxin-driven root bending is transmitted to promote development of the apical hook?
QZ Based on our work and published findings, we hypothesise that the initial gravity-stimulated, auxin-driven root bending might promote differences in mechanical forces at opposite sides of hypocotyl, which eventually would lead to an asymmetry in PIN expression. In support of such a scenario, it has been reported that changes in mechanical strains, such as modifications of turgor pressure or the application of external force, have considerable impact on subcellular trafficking and membrane localisation of PINs.
What do you think ABA and GA signalling are doing in the process?
MG Several recent studies including our work have reported that ABA affects the expression and membrane localisation of PIN auxin efflux carriers, key players in the regulation of apical hook formation. Intriguingly, ABA has been found to control stability of Rho guanine nucleotide exchange factors (RopGEFs), well-established regulators of ROPs, a family of small GTPases implicated in various cellular processes including control of PIN polar membrane localisation. This potential mechanistic link would definitely be worth further investigation. Unlike ABA, which reduces levels of PINs, GA has been found to interfere with lytic degradation of PIN proteins and consequently to enhance their accumulation at plasma membrane. So a speculative and plausible scenario is that the dynamic balance between ABA and GA signalling during embryo maturation and the early phases of germination tightly controls the expression and plasma membrane localisation of PIN auxin efflux carriers, thus leading to proper hypocotyl growth and apical hook formation. How these two hormonal pathways mutually interact and which components of their respective pathways are specifically involved in apical hook formation requires further research.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
QZ The most exciting moment for me was when I realised that the root gravitropic response is important for apical hook formation, by carefully observing kinetics of newly forming hooks. And when I found that green embryos are unable to form apical hooks despite a normal root gravity response, I knew this had provided an exciting model to study the mechanisms behind its development and in particular communication between these two organs.
MG My ‘aha!’ moment of this study was when I realised that seedlings germinating from mature embryos on medium supplemented with ABA resembled those grown from green embryos (at certain concentration the root is still growing, whereas cells in the shoot are not able to elongate anymore). This nicely supported our hypothesis that seedlings developing from green embryos might not able to form apical hooks because of their high levels of ABA. Thus, to stimulate formation of the apical hooks in these seedlings we might only need to deplete the excess ABA!
And what about the flipside: any moments of frustration or despair?
QZ For me, the moments of frustration were at the very beginning when I started this project. I had to peel off the Arabidopsis seed coats and isolate the embryos: you know, it’s very difficult to do this even under the microscope. I am so happy that after a long time practicing, my hands do not shake and I can operate freely with the seeds.
MG In line with the previous question, I tried many things to stimulate the growth of green embryos and observe the formation of an apical hook (auxin, GA, Norflurazon, abamine, etc.). It was really frustrating to see no rescue of growth, even in the ABA synthesis mutants or using chemicals that are supposed to inhibit ABA synthesis. It was not until the last trial, in which we combined several treatments (abamine and GA together in the aba2-1 mutant background), that we finally succeeded and could observe growth and apical hook formation in seedlings germinating from green embryos.
So what next for you two after this paper?
QZ I established my own research group in China after I left the Benková lab. Currently, we are studying bamboo as a new research model. Bamboo is probably the fastest growing plant on earth but until now nobody knew why it grows so fast: our group is trying to reveal the molecular mechanisms behind this. The research experiences in Eva’s lab really contributed a lot to my current research, and all the projects are running smoothly. So far, so good.
MG Next paper! We always have the next project in mind, and I am currently finishing experiments for what will be the next manuscript.
Where will this work take the Benková lab?
EB Now we know that if we want to fully understand mechanisms underlying formation of the apical hook, we have to also take into consideration processes occurring well before the bending of the hook itself, such as embryo maturation, outgrowth of the embryonic root and its alignment with the gravity vector. There are also many questions related to the cross-talk between ABA and GA and role of these hormones in coordination of the polar auxin transport required for proper formation of the apical hook. Finally, yet importantly, we still know very little about the pathways downstream of auxin that control the differential cell growth essential for hook development, but also bending of roots, hypocotyls or stems during gravi- or phototropism.
Finally, let’s move outside the lab – what do you like to do in your spare time?
QZ I left Vienna 4 years ago, and this question makes me recall the wonderful time I had in Austria, when my wife, my daughter and I would spend most of our free time traveling Vienna and the small towns around. We really enjoyed the charm of the music capital, and were impressed by the culture and atmosphere there.
MG I enjoy doing sport a lot, and the IST campus is a really good place for that. So I enjoy playing football and volleyball with my colleagues in the yards we have in the campus, and I also enjoy cycling and running in the woods around the campus. I feel privileged to have so many green areas surrounding the campus: there is nothing better for the brain than freshly produced oxygen from the woods!
EB I love gardening, not only because it is nice to have fresh vegetables and fruits, but I very much enjoy observing plants grow, seeing how they change from day to day, getting new leaves, flowers, tasty fruits… I also like travelling with my family, and especially hikes in mountains; a beautiful view into valley after a long tour is always worthwhile.


 (No Ratings Yet)
(No Ratings Yet)










 (8 votes)
(8 votes)