Closing Date: 15 March 2021
Post-doc in wing pattern formation and morphogenesis:
1.Job/ project description:
The research will involve using and refining an existing mathematical model of wing morphogenesis to explore whether it can be used to predict how wing morphology changes over generations in an artificial selection experiment. These predictions would be contrasted with predictions stemming from a quantitative genetics analysis of fly populations.
The research will take place in the Center of Excellence in Experimental and computational developmental biology of the Biotechnology Institute of the University of Helsinki, Finland.
For a full description of the project check the modeling in part of this funded project: http://www.biocenter.helsinki.fi/salazar/SalazarCiudad_research_plan.pdf
The job is for 1,5 years.
2. Background:
Why organisms are the way they are?
Can we understand the processes by which complex organisms are build in each generation and how these evolved?
The process of embryonic development is now widely acknowledged to be crucial to understand evolution since any change in the phenotype in evolution (e.g. morphology) is first a change in the developmental process by which this phenotype is produced. Over the years we have come to learn that there is a set of developmental rules that determine which phenotypic variation can possibly arise in populations due to genetic mutation (the so called genotype-phenotype map). Since natural selection can act only on existing phenotypic variation, these rules of development have an effect on the direction of evolutionary change.
Our group is devoted to understand these developmental rules and how these can help to better understand the direction of evolutionary change. The ultimate goal is to modify evolutionary theory by considering not only natural selection in populations but also developmental biology in populations. For that aim we combine mathematical models of embryonic development that relate genetic variation to morphological variation with population models. The former models are based on what is currently known in developmental biology.
There are two traditional approaches to study phenotypic evolution. One is quantitative genetics and one is developmental evolutionary biology. The former is based in the statistics of the association between genetic relatedness and phenotypic variation between individuals in populations, the latter in the genetic and bio-mechanical manipulation of the development of lab individuals. While the former models trait variation with an statistical linear approach the latter models it by deterministic non-linear models of gene networks and tissue bio-mechanics. For the most, these two approaches are largely isolated from each other.
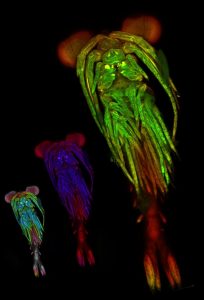
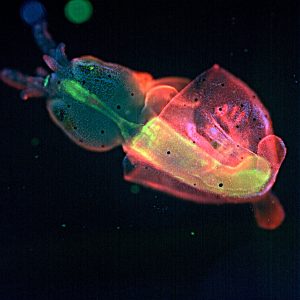
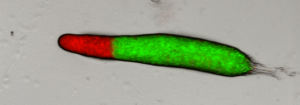
The current project aims to contrast and put together these two approaches in a specific easy to study system: the fly wing. In brief, we are growing fly populations and, in each generation, we select the founders of the next generation based on how close they resemble an arbitrary optimal morphology in their wings (based on the proportions between several of their traits). In each generation also, we estimate the G matrix and the selection gradient to see how well one can predict evolution in the next generation. The quantitative genetics predictions will be contrasted with the predictions stemming from a wing morphogenesis model that we built based on our current understanding of wing developmental biology (see Dev Cell. 2015 Aug 10;34(3):310-22 for the model and for slightly similar approaches: Nature. 2013 May 16;497(7449):361-4. and Nature. 2010 Mar 25;464(7288):583-6).
Our center of excellence includes groups working in tooth, wing, hair and mammary glands development. In addition to evolutionary and developmental biologists the center of excellence includes bioinformaticians, populational and quantitative geneticists, systems biologists and paleontologists. The group leaders of the center involved in this project are Jukka Jernvall, Salazar-Ciudad and Shimmi.
“The Academy of Finland’s Centres of Excellence are the flagships of Finnish research. They are close to or at the very cutting edge of science in their fields, carving out new avenues for research, developing creative research environments and training new talented researchers for the Finnish research system.”
3. Requirements:
The applicant must hold a PhD in either evolutionary biology, developmental biology or, preferably, in evolutionary developmental biology (evo-devo). Applicants with a PhD in theoretical or mathematical biology are also welcome.
Programming skills or a willingness to acquire them is required.
The most important requirement is a strong interest and motivation on science and evolution. A capacity for creative and critical thinking is also required.
4. Description of the position:
The fellowship will be for a period of up to 1,5 years (100% research work: no teaching involved).
Salary according to Finnish postdoc salaries.
5. The application must include:
-Motivation letter including a statement of interests
-CV (summarizing degrees obtained, subjects included in degree and grades, average grade).
-Summary of PhD project, its main conclusions and its underlying motivation.
-Application should be sent to: isaac.salazar@helsinki.fi
No official documents are required for the application first stage but these may be required latter on.
6. Deadline:
There is no specific deadline but the position will start in July 2019.
7. Interested candidates should check the centres page:
http://www.biocenter.helsinki.fi/bi/evodevo/ECDev.html
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (2 votes)
(2 votes)