February in preprints
Posted by the Node, on 1 March 2018
Our latest monthly trawl for developmental biology (and other cool) preprints. Let us know if we missed anything.
On February 20th, The Company of Biologists launched preLights, a community-led preprint highlighting service. A panel of early career researchers (the ‘preLighters’) select and comment on recent preprints that caught their eye, and encourage preprint authors to answer any questions about the work that they had. So far it looks great, and the developmental biology content has been especially good (see the dedicated subject category). We’d love to know what you think: you can contact the team via the site or the Twitter feed.
The idea behind the site was influenced in part by this list – as it got longer and longer (reflecting increased preprint usage), we were wondering how else we could encourage and promote the discussion of preprints, and the preLights idea took form. Rest assured that this list will live on, at least until the point at which it gets impossibly long!
And here’s the list – all the developmental biology I could find, plus relevant and cool other preprints thrown in for good measure.
The preprints were hosted on bioRxiv, PeerJ, and arXiv. Use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice & education
Why not…
Developmental biology
| Patterning & signalling
Human axial progenitors generate trunk neural crest cells. Thomas J.R. Frith, Ilaria Granata, Erin Stout, Matthew Wind, Oliver Thompson, Katrin Neumann, Dylan Stavish, Paul R Heath, James O.S. Hackland, Konstantinos Anastassiadis, Mina Gouti, James Briscoe, Valerie Wilson, Mario R Guarracino, Peter W Andrews, Anestis Tsakiridis
Lineage tracing axial progenitors using Nkx1.2CreERT2 mice defines their trunk and tail contributions. Aida Rodrigo Albors, Pamela A Halley, Kate G Storey
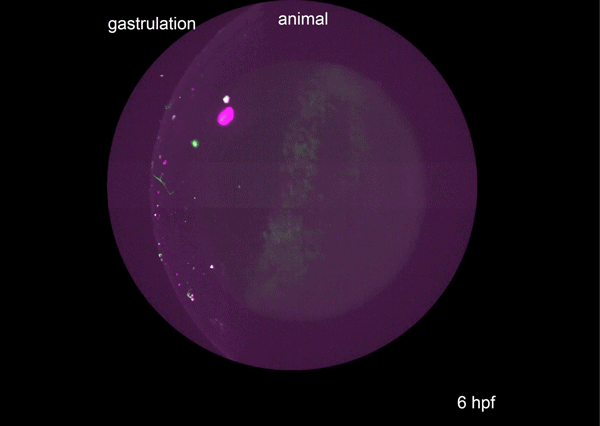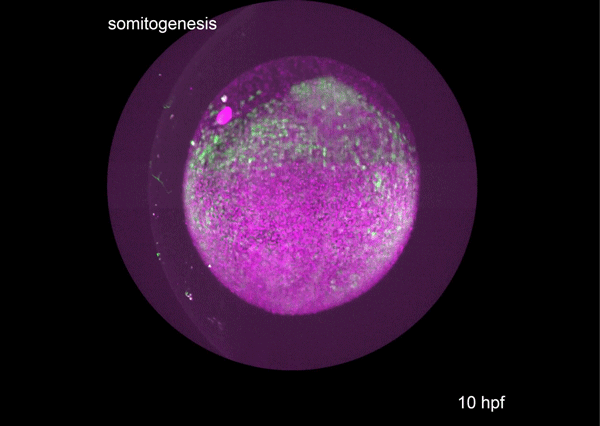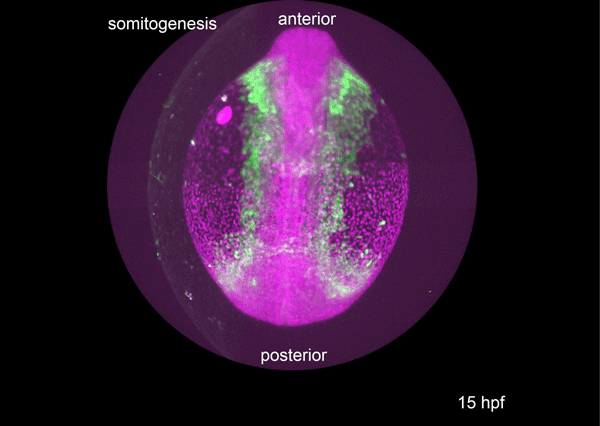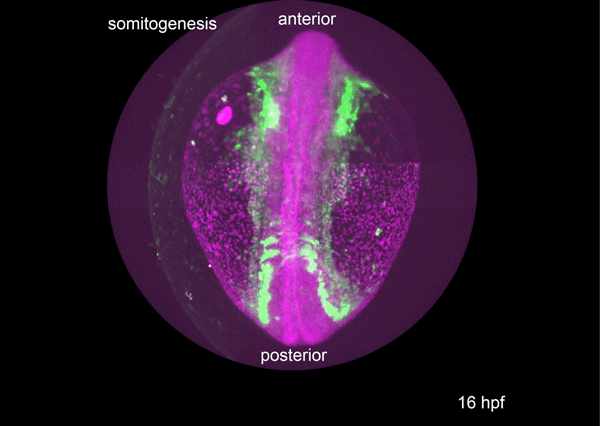
A conserved regulatory program drives emergence of the lateral plate mesoderm. Christopher Hess, Karin Dorien Prummel, Susan Nieuwenhuize, Hugo Parker, Katherine W. Rogers, Iryna Kozmikova, Claudia Racioppi, Sibylle Burger, Eline C. Brombacher, Alexa Burger, Anastasia Felker, Elena Chiavacci, Gopi Shah, Jan Huisken, Zbynek Kozmik, Lionel Christiaen, Patrick Mueller, Marianne Bronner, Robb Krumlauf, Christian Mosimann
Hedgehog signaling controls progenitor differentiation timing during heart development. Megan Rowton, Andrew D. Hoffmann, Jeffrey D. Steimle, Xinan Holly Yang, Alexander Guzzetta, Sonja Lazarevic, Chul Kim, Nikita Deng, Emery Lu, Jessica Jacobs-Li, Shuhan Yu, Erika Hanson, Carlos Perez-Cervantes, Sunny Sun-Kin Chan, Kohta Ikegami, Daniel J. Garry, Michael Kyba, Ivan P. Moskowitz
SOX2 Regulation by Hedgehog Signaling Controls Adult Lingual Epithelium Homeostasis. David Castillo-Azofeifa, Kerstin Seidel, Lauren Gross, Belkis Jacquez, Ophir D Klein, Linda A. Barlow
Extracellular Pax6 regulates tangential Cajal-Retzius cell migration. Hadhemi Kaddour, Eva Coppola, Ariel Di Nardo, Andrea Wizenmann, Michel Volovitch, Alain Prochiantz, Alessandra Pierani
Axon Guidance Signaling Modulates Epithelial to Mesenchymal Transition in Stem Cell-Derived Retinal Pigment Epithelium. Srinivas R. Sripathi, Melissa M. Liu, Ming-Wen Hu, Jun Wan, Jie Cheng, Yukan Duan, Joseph Mertz, Karl Wahlin, Julien Maruotti, Cynthia A Berlinicke, Jiang Qian, Donald J. Zack
Neuroligins mediate presynaptic maturation through brain-derived neurotrophic factor signaling. Andoniya Petkova, Nina Goedecke, Martin Korte, Thomas Dresbach
In vivo clonal analysis reveals spatiotemporal regulation of thalamic nucleogenesis. Samuel Wong, Earl Parker Scott, Wenhui Mu, Xize Guo, Ella Borgenheimer, Madeline Freeman, Guo-li Ming, Qing-Feng Wu, Hongjun Song, Yasushi Nakagawa
Osteoclasts control sensory neurons axonal growth through epidermal growth factor receptor signaling. Estrela Neto, Daniela Sousa, Francisco Conceição, Luís Leitão, Cecília Juliana Alves, Inês Alencastre, Jonathan West, Richard O. C. Oreffo, Meriem Lamghari
FGF activity asymmetrically regulates the timing of habenular neurogenesis in a Nodal-dependent manner. Benjamin J Dean, Joshua T Gamse, Shu-Yu Wu
The transcription factor Tfap2e/AP-2ε plays a pivotal role in maintaining the identity of basal vomeronasal sensory neurons. Jennifer M Lin, Ed Zandro M Taroc, Jesus A Frias, Aparna Prasad, Allison N Catizone, Morgan A Sammons, Paolo E. Forni
Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Elke F Roovers, Lucas JT Kaaij, Stefan Redl, Alfred W Bronkhorst, Kay Wiebrands, Antonio Miguel de Jesus Domingues, Hsin-Yi Huang, Chung-Ting Han, Willi Salvenmoser, Dominic Gruen, Falk Butter, Alexander van Oudenaarden, Rene F Ketting
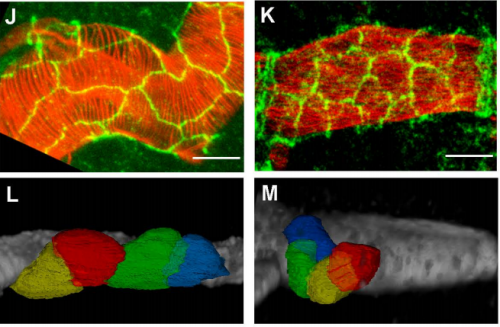
Blimp-1 mediates the temporal control of embryonic tube maturation. Arzu Ozturk-Colak, Camille Stephan-Otto Attolini, Jordi Casanova, Sofia J. Araujo
The integrated stress response regulates BMP signaling through effects on translation. Elke Malzer, Caia S Dominicus, Joseph E Chambers, Souradip Mookerjee, Stefan J Marciniak
Dietary lipids modulate Notch signaling and influence adult intestinal development and metabolism in Drosophila. Rebecca Obniski, Matthew Sieber, Allan C Spradling
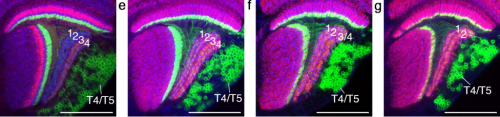
Spatio-temporal relays control layer specificity of motion-direction sensitive neurons in Drosophila. Holger Apitz, Iris Salecker
Torso-like is a component of the hemolymph and regulates the insulin signalling pathway in Drosophila. Michelle A Henstridge, Lucinda Aulsebrook, Takashi Koyama, Travis K Johnson, James C Whisstock, Tony Tiganis, Christen K Mirth, Coral G Warr
Variability in timing of a β-catenin pulse biases a stochastic cell fate decision in C. elegans. Jason R Kroll, Jasonas Tsiaxiras, Jeroen S van Zon
The Atypical Rho GTPase CHW-1 Works With SAX-3/Robo to Mediate Axon Guidance in Caenorhabditis elegans. Jamie Alan, Sara Robinson, Katie Magsig, Rafael Demarco, Erik A. Lundquist
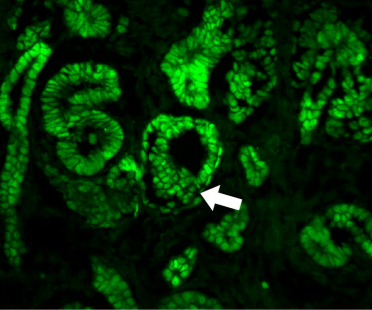
Phospho-regulation of Atoh1 is required for plasticity of secretory progenitors and tissue regeneration. Goran Tomic, Edward Morrissey, Sarah Kozar, Shani Ben-Moshe, Alice Hoyle, Roberta Azzarelli, Richard Kemp, Chandra Sekhar Reddy Chilamakuri, Shalev Itzkovitz, Anna Philpott, Douglas Winton
The TMEM16A Channel Mediates the fast polyspermy block in Xenopus Laevis. Katherine L Wozniak, Wesley A Phelps, Maiwase Tembo, Miler T Lee, Anne E Carlson
Early lethality of embryos derived from transgenic Xenopus females is associated with reduced ovarian grem1 expression. Caroline W Beck, Joanna Ward, Lisa Troise, Catherine Brochard
Genetic inactivation of alpha-synuclein affects embryonic development of dopaminergic neurons of the substantia nigra but not the ventral tegmental area in mouse brain. Tatyana V Tarasova, Olga A Lytkina, Valeria V Goloborshcheva, Larisa N Skuratovskaya, Alexandr I Antohin, Ruslan K Ovchinnikov, Michail S Kukharsky
Single-cell analysis of progenitor cell dynamics and lineage specification of the human fetal kidney. Rajasree Menon, Edgar A. Otto, Austin Kokoruda, Jian Zhou, Zidong Zhang, Euisik Yoon, Yu-Chih Chen, Olga Troyanscaya, Jason Spence, Matthias Kretzler, Cristina Cebrian
A hypomorphic Stip1 allele reveals the requirement for chaperone networks in mouse development and aging. Rachel Lackie, Marilene H Lopes, Sali M.K. Farhan, Abdul Razzaq, Gilli Moshitzky, Mariana B Prado, Flavio H Beraldo, Andrzej Maciejewski, Robert Gros, Jue Fan, Wing-Yiu Choy, David Greenberg, Vilma R Martins, Martin Duennwald, Hermona Soreq, Vania F Prado, Marco A. M. Prado
Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Jacob Daane, Jennifer Lanni, Ina Rothenberg, Guiscard Seebohm, Charles Higdon, Stephen Johnson, Matthew Harris
GEMC1 and CCNO are required for efferent duct development and male fertility. Berta Terre, Michael Lewis, Gabriel Gil-Gomez, Travis H Stracker
| Morphogenesis & mechanics
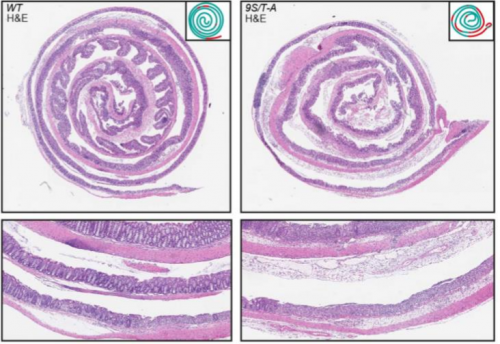
Characterization of convergent thickening, a major convergence force producing morphogenic movement in amphibians. David R Shook, Jason Wen, Ana Rolo, Brian Francica, Destiny Dobins, Paul Skoglund, Douglas DeSimone, Rudolf Winklbauer, Ray Keller
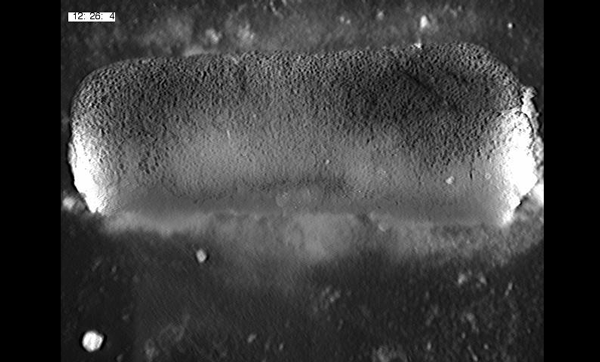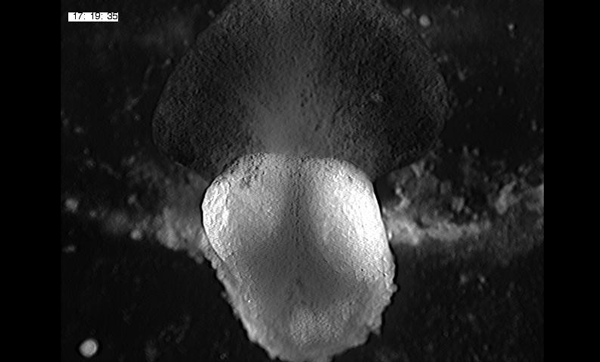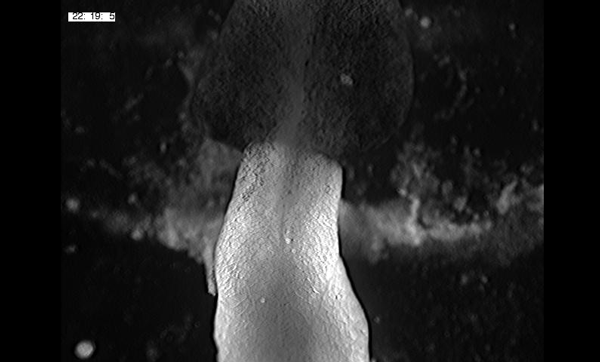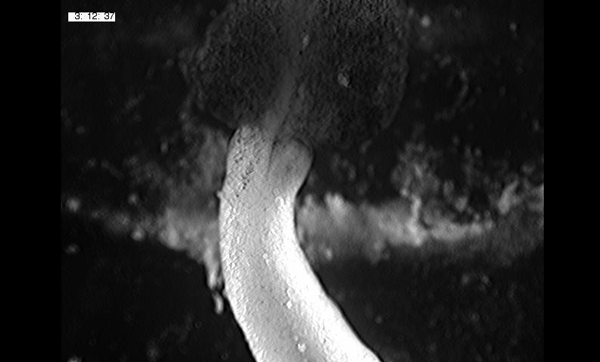
Large, long range tensile forces drive convergence during Xenopus blastopore closure and body axis elongation. David R Shook, Ray Keller, Lance Davidson, Eric M. Kasprowicz
Contact-dependent cell communications drive morphological invariance during ascidian embryogenesis. Leo Guignard, Ulla-Maj Fiuza, Bruno Leggio, Emmanuel Faure, Julien Laussu, Lars Hufnagel, Gregoire Malandain, Christophe Godin, Patrick Lemaire
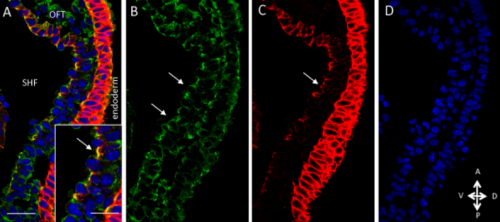
Tbx1 regulates extracellular matrix- and cell-cell interactions in the second heart field. Antonio Baldini, Daniela Alfano, Alessandra Altomonte, Marchesa Bilio
An unconventional myosin, myosin 1d regulates Kupffer’s vesicle morphogenesis and laterality. Manush Saydmohammed, Hisato Yagi, Michael Calderon, Madeline J Clark, Timothy Feinstein, Ming Sun, Donna B Stolz, Simon C Watkins, Jeffrey Amack, Cecilia Lo, Michael Tsang
Myosin1D is an evolutionarily conserved determinant of animal Left/Right asymmetry. Thomas Juan, Charles Géminard, Jean-Baptiste Coutelis, Delphine Cerezo, Sophie Polès, Stéphane Noselli, Maximilian Fürthauer
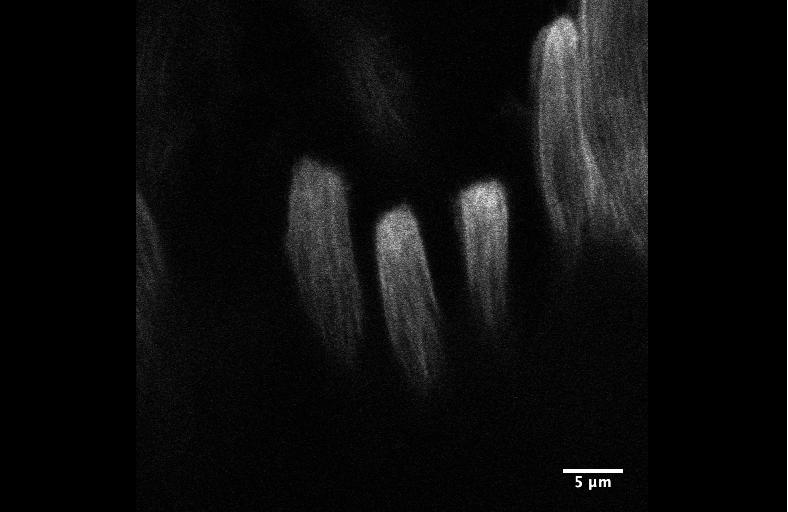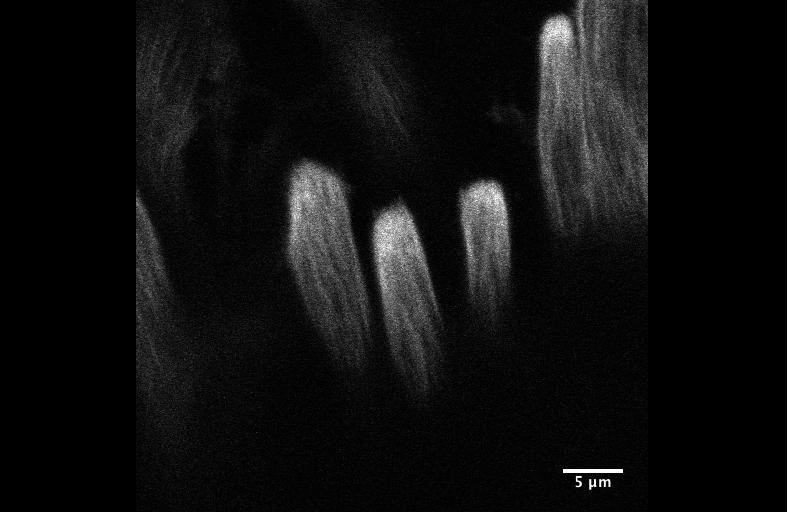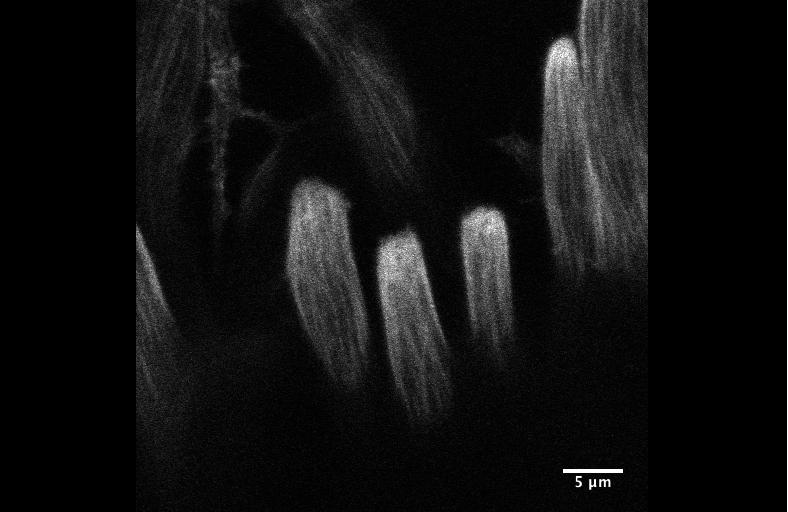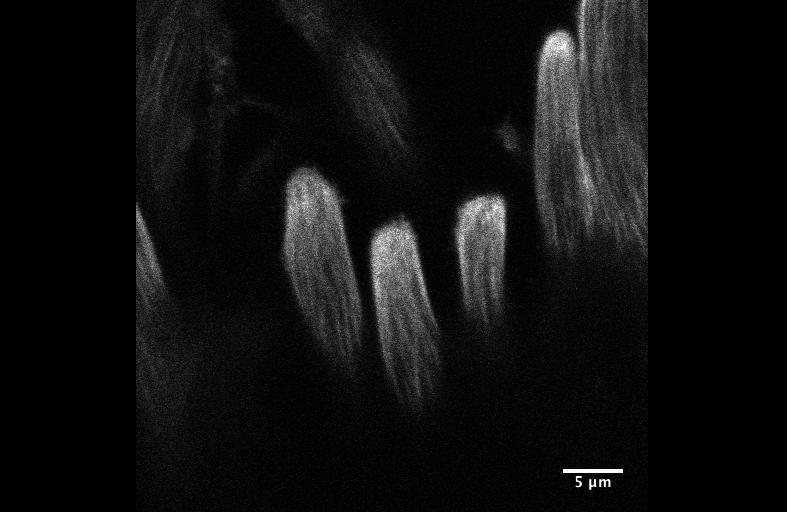
Integrin signaling downregulates filopodia in muscle-tendon attachment. Benjamin Richier, Yoshiko Inoue, Ulrich Dobramysl, Jonathan Friedlander, Nicholas H. Brown, Jennifer Louise Gallop
Long-range Notch-mediated tissue patterning requires actomyosin contractility. Ginger Hunter, Li He, Norbert Perrimon, Guillaume Charras, Edward Giniger, Buzz Baum
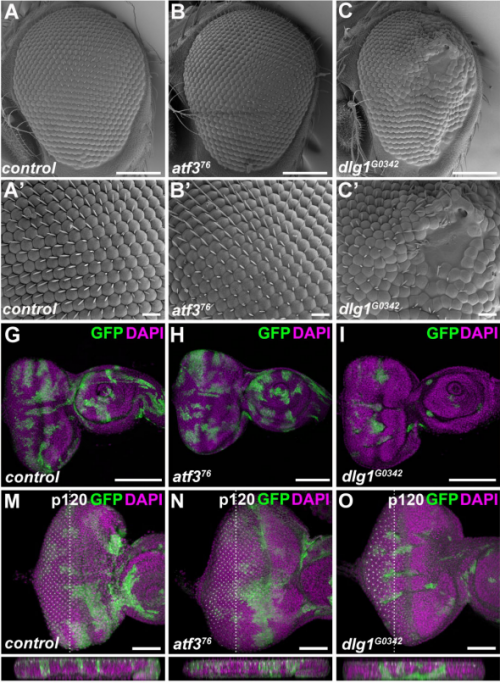
Atf3 links loss of epithelial polarity to defects in cell differentiation and cytoarchitecture. Colin D. Donohoe, Gabor Csordas, Andreia Correia, Marek Jindra, Corinna Klein, Bianca Habermann, Mirka Uhlirova
Myosin Heavy Chain-embryonic is a crucial regulator of skeletal muscle development and differentiation. Akashi Sharma, Megha Agarwal, Amit Kumar, Pankaj Kumar, Masum Saini, Gabrielle Kardon, Sam Mathew
The C. elegans heterochronic gene lin-28 coordinates the timing of hypodermal and somatic gonadal programs for hermaphrodite reproductive system morphogenesis. Sungwook Choi, Victor Ambros
Non-canonical apical constriction shapes emergent matrices in C. elegans. Sophie S Katz, Chloe Maybrun, Alison Renee Frand
A microtubule-based mechanism predicts cell division orientation in plant embryogenesis. Bandan Chakrabortty, Viola Willemsen, Thijs de Zeeuw, Che-Yang Liao, Dolf Weijers, Bela Mulder, Ben Scheres

Feeling the force: how pollen tubes deal with obstacles. Jan T Burri, Hannes Vogler, Nino F Läubli, Chengzhi Hu, Ueli Grossniklaus, Bradley J Nelson
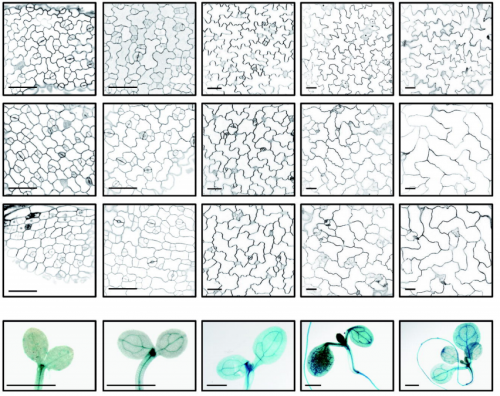
Microtubule-associated protein IQ67 DOMAIN5 regulates interdigitation of leaf pavement cells in Arabidopsis thaliana. Dipannita Mitra, Pratibha Kumari, Jakob Quegwer, Sandra Klemm, Birgit Moeller, Yvonne Poeschl, Paul Pflug, Gina Stamm, Steffen Abel, Katharina Bürstenbinder
Cell death in cells overlying lateral root primordia contributes to organ growth in Arabidopsis. Sacha Escamez, Benjamin Bollhoner, Hardy Hall, Domenique Andre, Beatrice Berthet, Ute Voss, Amnon Lers, Alexis Maizel, Malcolm Bennett, Hannele Tuominen
FERONIA’s sensing of cell wall pectin activates ROP GTPase signaling in Arabidopsis. Wenwei Lin, Wenxin Tang, Charles Anderson, Zhenbiao Yang
Adaptive diversification of growth allometry in the plant Arabidopsis thaliana. Francois Vasseur, Moises Exposito-Alonso, Oscar Javier Ayala-Garay, George Wang, Brian J Enquist, Denis Vile, Cyrille Violle, Detlef Weigel
A mechanism for the proliferative control of tissue mechanics in the absence of growth. Min Wu, Madhav Mani
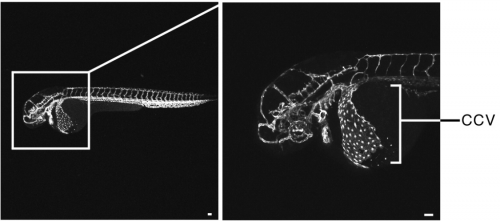
Endothelia extrude apoptotic cells to maintain a constant barrier. Tara Mleynek, Dean Li, Jody Rosenblatt, Michael Jarman Redd, Aubrey Chan, Yapeng Gu
Cellular Migration May Exhibit Intrinsic Left-Right Asymmetries: A meta-analysis. Kelly G. Sullivan, Laura N. Vandenberg, Michael Levin
Increased prenatal brain growth in a transgenic mouse model decreases cranial development: An expensive tissue hypothesis for the skull. Elisabeth K N López, Matthew J Ravosa
In vivo optochemical control of cell contractility at single cell resolution by Ca2+ induced myosin activation. Deqing Kong, Fred Wolf, Joerg Grosshans.
A quantitative study of blood circulation in the developing adult ascidian tunicate Ciona savignyi (Cionidae). Michael W Konrad
Tissue-specific activities of the Fat1 cadherin cooperate to control neuromuscular morphogenesis. Francoise Helmbacher
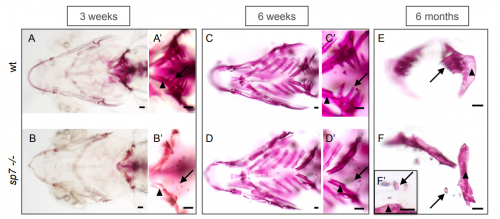
Zebrafish sp7 mutants show tooth cycling independent of attachment, eruption and poor differentiation of teeth. Erika Kague, Paul Eckhard Witten, Mieke Soenens, Camila Lovaglio Campos, Tiago Lubiana, Shannon Fisher, Chrissy Hammond, Kate Robson Brown, Maria Rita Passos Bueno, Ann Huysseune
| Genes & genomes
Disruption in A-to-I editing levels affects C. elegans development more than a complete lack of editing. Nabeel S. Ganem, Noa Ben-Asher, Aidan C. Manning, Sarah N. Deffit, Michael C. Washburn, Emily C. Wheeler, Gene W Yeo, Orna Ben-Naim Zgayer, Einav Mantsur, Heather A. Hundley, Ayelet T Lamm
ADAR1 A-to-I RNA editing alters codon usage. Pavla Brachova, Nehemiah S Alvarez, Xiaoman Hong, Kailey A. Vincent, Keith E. Latham, Lane K. Christenson
Identification of piRNA binding sites reveals the Argonaute regulatory landscape of the C. elegans germline. En-Zhi Shen, Hao Chen, Ahmet R. Ozturk, Shikui Tu, Masaki Shirayama, Wen Tang, Yue-He Ding, Si-Yuan Dai, Zhiping Weng, Craig C. Mello
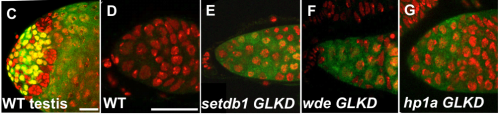
H3K9 methylation maintains female identity in Drosophila germ cells through repression of the spermatogenesis program. Anne Smolko, Laura Shapiro-Kulnane, Helen Salz
Fanconi Anemia FANCM/FNCM-1 and FANCD2/FCD-2 are required for maintaining histone methylation levels and interact with the histone demethylase LSD1/SPR-5 in C. elegans. Hyun-Min Kim, Sara E Beese-Sims, Monica P. Colaiácovo
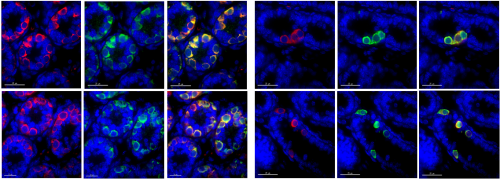
GTSF-1 is required for the formation of a functional RNA-dependent RNA Polymerase complex in C. elegans. Miguel V. Almeida, Sabrina Dietz, Stefan Redl, Emil Karaulanov, Andrea Hildebrandt, Christian Renz, Helle D. Ulrich, Julian König, Falk Butter, René F. Ketting
Dissection of the microProtein miP1 floral repressor complex in Arabidopsis. Vandasue L. Rodrigues, Ulla Dolde, Daniel Straub, Tenai Eguen, Esther Botterweg-Paredes, Bin Sun, Shinyoung Hong, Moritz Graeff, Man-Wah Li, Joshua Gendron, Stephan Wenkel
Differences in firing efficiency, chromatin and transcription underlie the developmental plasticity of Arabidopsis originome. Joana Sequeira-Mendes, Zaida Vergara, Ramon Peiro, Jordi Morata, Irene Araguez, Celina Costas, Raul Mendez-Giraldez, Josep Casacuberta, Ugo Bastolla, Crisanto Gutierrez
Incomplete reprogramming of cell-specific epigenetic marks during asexual reproduction leads to heritable phenotypic variation in plants. Anjar Wibowo, Claude Becker, Julius Durr, Jonathan Price, Stijn Staepen, Sally Hilton, Hadi Putra, Ranjith Papareddy, Quentin Saintain, Sarah Harvey, Gary D Bending, Paul Schulze-Lefert, Detlef Weigel, Jose Gutierrez-Marcos
Epigenomic plasticity of Arabidopsis msh1 mutants under prolonged cold stress. Sunil Kumar Kenchanmane Raju, Mon-Ray Shao, Yashitola Wamboldt, Sally Mackenzie
A Lotus japonicus E3 ligase interacts with the Nod factor receptor 5 and positively regulates nodulation. Daniela Tsikou, Estrella E. Ramirez, Ioanna S Psarrakou, Jaslyn E Wong, Dorthe B Jensen, Erika Isono, Simona Radutoiu, Kalliope K Papadopoulou
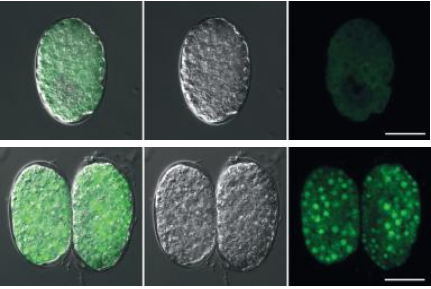
Dazl regulates germ cell survival through a network of polyA proximal mRNA interactions. Leah L Zagore, Thomas J Sweet, Molly M Hannigan, Sebastien M Weyn-Vanhentenryck, Chaolin Zhang, Donny D Licatalosi
Enhancer RNAs are necessary and sufficient for activity-dependent neuronal gene transcription. Nancy V.N. Gallus, Rhiana A. Simon, Aaron J Salisbury, Jasmin S. Revanna, Kendra D. Bunner, Katherine E. Savell, Faraz A. Sultan, Charles A. Gersbach, Jeremy J. Day
Mef-2 and p300 interact to regulate expression of the homeostatic regulator Pumilio in Drosophila. Wei-Hsiang Lin, Richard Baines
APC/C-Vihar regulates centrosome activity and stability in the Drosophila germline. Alexis L. Braun, Francesco Meghini, Gema Villa-Fombuena, Elisa M. Fernandez, Morgane Guermont, David M Glover, Maria D. Martin-Bermudo, Acaimo Gonzalez-Reyes, Yuu Kimata
A Genetic Program Mediates Cold-warming Response and Promotes Stress-induced Phenoptosis in C. elegans. Wei Jiang, Yuehua Wei, Yong Long, Arthur Owen, Bingying Wang, Xuebing Wu, Shuo Luo, Yongjun Dang, Dengke K. Ma
DNA methylation dynamics during embryonic development and postnatal maturation of the mouse auditory organ of Corti. Ofer Yizhar-Barnea, Cristina Valensisi, Kamal Kishore, Naresh Doni Jayavelu, Colin Andrus, Tal Koffler-Brill, Kathy Ushakov, Kobi Perl, Yael Noy, Yoni Bhonker, Mattia Pelizzola, R. David Hawkins, Karen B. Avraham
Aggregation recovers developmental plasticity in mouse polyploid embryos. Hiroyuki Imai, Wataru Fujii, Ken Takeshi Kusakabe, Yasuo Kiso, Kiyoshi Kano
Capturing the onset of PRC2-mediated repressive domain formation. Ozgur Oksuz, Varun Narendra, Chul-Hwan Lee, Nicolas Descostes, Gary LeRoy, Ramya Raviram, Lili Blumenberg, Kelly Karch, Pedro R. Rocha, Benjamin A. Garcia, Jane A. Skok, Danny Reinberg
Cell type-dependent control of p53 transcription and enhancer activity by p63. Gizem Karsli Uzunbas, Faraz Ahmed, Morgan A Sammons
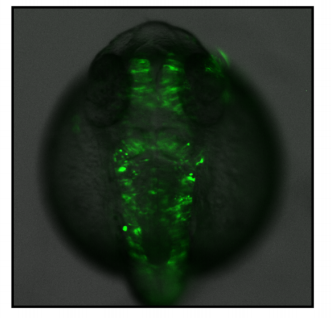
Genome-wide enhancer – gene regulatory maps in two vertebrate genomes. Yves Clément, Patrick Torbey, Pascale Gilardi-Hebenstreit, Hugues Roest Crollius
KLF6 and STAT3 co-occupy regulatory DNA and functionally synergize to promote axon growth in CNS neurons. Zimei Wang, Ishwariya Venkatesh, Vatsal Mehra, Matthew Simpson, Brian Maunze, Erik Eastwood, Lyndsey Holan, Murray Blackmore
Germline DNA replication timing shapes mammalian genome composition. Yishai Yehida, Britny Blumenfeld, Nina Mayorek, Kirill Makedonski, Oriya Vardi, Yousef Mansour, Hagit Masika, Marganit Farago, Shulamit Baror-Sebban, Yosef Buganim, Amnon Koren, Itamar Simon
| Stem cells, regeneration & disease modelling
Tracing the Transitions from Pluripotency to Germ Cell Fate with CRISPR Screening. Jamie A Hackett, Yun Huang, Ufuk Gunesdogan, Kristjan Holm-Gretarsson, Toshihiro Kobayashi, Azim M Surani
Neutralizing Gatad2a-Chd4-Mbd3 Axis within the NuRD Complex Facilitates Deterministic Induction of Naive Pluripotency. Nofar Mor, Yoach Rais, Shani Peles, Daoud Sheban, Alejandro Aguilera-Castrejon, Asaf Zviran, Dalia Elinger, Sergey Viukov, Shay Geula, Vladislav Krupalnik, Mirie Zerbib, Elad Chomsky, Lior Lasman, Tom Shani, Jonathan Bayerl, Ohad Gafni, Suhair Hanna, Jason Buenrostro, Tzachi Hagai, Hagit Masika, Yehudit Bergman, William J. Greenleaf, Miguel A. Esteban, Yishai Levin, Rada Massarwa, Yifat Merbl, Noa Novershtern, Jacob H. Hanna
Delayed activation of the DNA replication licensing system in Lgr5(+) intestinal stem cells. Thomas Carroll, Ian Newton, Yu Chen, Julian J. Blow, Inke Nathke
Efficient differentiation of vascular endothelial cells from dermal-derived mesenchymal stem cells induced by endothelial cell lines conditioned medium. Ling Zhou, Xuping Niu, Jiannan Liang, Junqin Li, Jiao Li, Yueai Cheng, Yanfeng Meng, Qiang Wang, Xiaoli Yang, Gang Wang, Yu Shi, Erle Dang, Kaiming Zhang
Clonal Level Lineage Commitment Pathways of Hematopoietic Stem Cells In Vivo. Rong Lu, Agnieszka Czechowicz, Jun Seita, Du Jiang, Irving L Weissman
Hematopoietic Stem And Progenitor Cells Use Podosomes To Transcellularly Home To The Bone Marrow. Timo Rademakers, Marieke Goedhart, Mark Hoogenboezem, Alexander García Ponce, Jos van Rijssel, Maryna Samus, Michael Schnoor, Stefan Butz, Stephan Huveneers, Dietmar Vestweber, Martijn A Nolte, Carlijn Voermans, Jaap D van Buul
Epigenetic aging of human hematopoietic cells is not accelerated upon transplantation into mice. Joana Frobel, Susann Rahmig, Julia Franzen, Claudia Waskow, Wolfgang Wagner
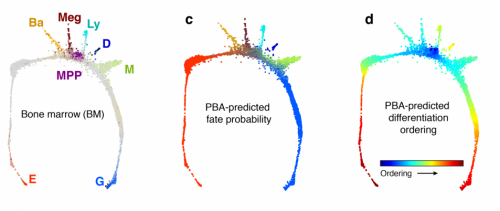
Emergence of the erythroid lineage from multipotent hematopoiesis. Betsabeh Khoramian Tusi, Samuel L Wolock, Caleb Weinreb, Yung Hwang, Daniel Hidalgo, Rapolas Zilionis, Ari Waisman, Jun Huh, Allon M Klein, Merav Socolovsky
An immortalised mesenchymal stem cell line maintains mechano-responsive behaviour and can be used as a reporter of substrate stiffness. Asier Galarza Torre, Joshua E Shaw, Amber Wood, Hamish T J Gilbert, Oana Dobre, Paul Genever, Keith Brennan, Stephen M Richardson, Joe Swift
Heat shock induces the depletion of Oct4 in mouse blastocysts and stem cells. Mo-bin Cheng, Xue Wang, Yue Huang, Ye Zhang
Single-cell transcriptome of antler stem cells from antlerogenic periosteum. Hengxing Ba, Datao Wang, Weiyao Wu, Hongmei Sun, Chunyi Li
Xist Intron 1 Repression by TALE Transcriptional Factor Improves Somatic Cell Reprogamming in Mice. Jindun Zhang, Xuefei Gao, Jian Yang, Xiaoying Fan, Wei Wang, Yanfeng Liang, Lihong Fan, Hongmei Han, Xiaorong Xu, Fuchou Tang, Siqin Bao, Pentao Liu, Xihe Li
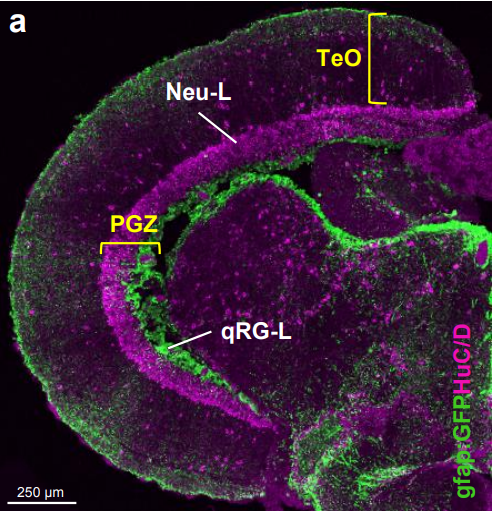
Tectal stem cells display diverse regenerative capacities. Benjamin W Lindsey, Georgia E Aitken, Jean K Tang, Mitra Khabooshan, Celia Vandestadt, Jan Kaslin
Developmental chromatin restriction of pro-growth gene networks acts as an epigenetic barrier to axon regeneration in cortical neurons. Ishwariya Venkatesh, Vatsal Mehra, Zimei Wang, Ben Califf, Murray Blackmore
Neuregulin-1 exerts molecular control over axolotl lung regeneration through ErbB family receptors. Tyler B Jensen, Peter Giunta, Natalie Grace Schulz, Yaa Kyeremateng, Hilary Wong, Adeleso Adesina, James R Monaghan
Targeting TrkB with a brain-derived neurotrophic factor mimetic promotes myelin repair in the brain. Jessica Louise Fletcher, Rhiannon J Wood, Jacqueline Nguyen, Eleanor ML Normon, Christine MK Jun, Alexa R Prawdiuk, Melissa Biemond, Huynh TH Nguyen, Susan E Northfield, Richard A Hughes, David G Gonsalvez, Junhua Xiao, Simon S Murray
Age-dependent dormant resident progenitors are stimulated by injury to regenerate Purkinje neurons. Nermin Sumru Bayin, Alexandre Wojcinski, Aurelien Mourton, Hiromitsu Saito, Noboru Suzuki, Alexandra L Joyner
Reversing Glial Scar Back To Neural Tissue Through NeuroD1-Mediated Astrocyte-To-Neuron Conversion. Lei Zhang, Zhuofan Lei, Ziyuan Guo, Zifei Pei, Yuchen Chen, Fengyu Zhang, Alice Cai, Yung Kin Mok, Grace Lee, Vishal Swaminathan, Fan Wang, Yuting Bai, Gong Chen
Loss of embryonic neural crest cardiomyocytes causes adult hypertrophic cardiomyopathy. Sarah Abdul-Wajid, Bradley Demarest, H. Joseph Yost
p53 deletion rescues apoptosis and microcephaly in a Kif20b mouse mutant. Jessica Neville Little, Noelle D. Dwyer
Altered neocortical gene expression, brain overgrowth and functional over-connectivity in Chd8 haploinsufficient mice. Philipp Suetterlin, Shaun Hurley, Conor Mohan, Kimberley L. H. Riegman, Marco Pagani, Angela Caruso, Jacob Ellegood, Alberto Galbusera, Ivan Crespo-Enriquez, Caterina Michetti, Yohan Yee, Robert Ellingford, Olivier Brock, Alessio Delogu, Philippa Francis-West, Jason P. Lerch, Maria Luisa Scattoni, Alessandro Gozzi, Cathy Fernandes, Albert Basson
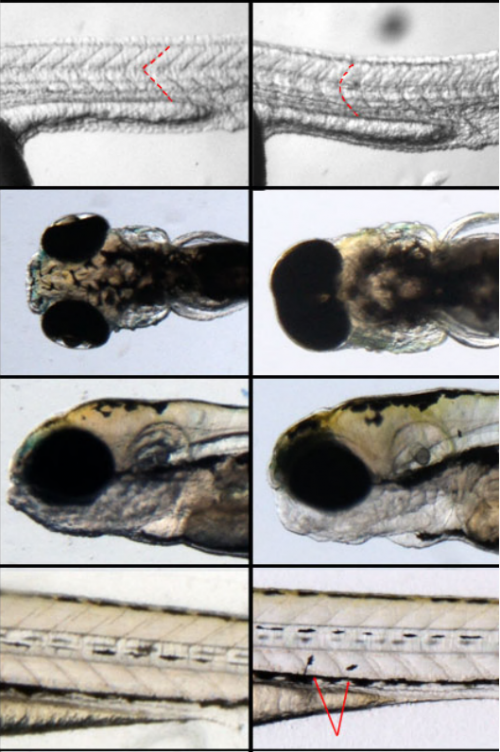
Survey of human chromosome 21 gene expression effects on early development in Danio rerio. Sarah Edie, Norann A Zaghloul, Carmen C Leitch, Donna K Klinedinst, Janette Lebron, Joey F Thole, Andrew S. McCallion, Nicholas Katsanis, Roger H. Reeves
Regulation of midgut cell proliferation impacts Aedes aegypti susceptibility to dengue virus. Mabel L. Taracena, Vanessa Bottino-Rojas, Octavio A.C. Talyuli, Ana Beatriz Walter-Nuno, Jose Henrique M. Oliveira, Yesseinia I. Anglero-Rodriguez, Michael B. Wells, George Dimopoulos, Pedro L. Oliveira, Gabriela O. Paiva-Silva
Ablation of Ezh2 in neural crest cells leads to Hirschsprung’s disease-like phenotype in mice. Joomyeong Kim, Hana Kim, Ingeborg M. Langohr, Mohammad Faisal, Margaret McNulty, Caitlin Thorn
Building a schizophrenia genetic network: Transcription Factor 4 regulates genes involved in neuronal development and schizophrenia risk. Hanzhang Xia, Fay M Jahr, Nak-Kyeong Kim, Linying Xie, Andrey A Shabalin, Julien Bryois, Douglas H Sweet, Mohamad M Kronfol, Preetha Palasuberniam, MaryPeace McRae, Brien P Riley, Patrick F Sullivan, Edwin J van den Oord, Joseph McClay
Pervasive epistasis modulates neurodevelopmental defects of the autism-associated 16p11.2 deletion. Janani Iyer, Mayanglambam Dhruba Singh, Matthew Jensen, Payal Patel, Lucilla Pizzo, Emily Huber, Haley Koerselman, Alexis T. Weiner, Paola Lepanto, Komal Vadodaria, Alexis Kubina, Qingyu Wang, Abigail Talbert, Sneha Yennawar, Jose Badano, J. Robert Manak, Melissa M. Rolls, Arjun Krishnan, Santhosh Girirajan
The critical role of ASD-related gene CNTNAP3 in regulating synaptic development and social behavior in mice. Dali Tong, Ruiguo Chen, Yulan Lu, Weike Li, Yuefang Zhang, Junkai Lin, Lingjie He, Ting Dang, Shifang Shan, Xiaohong Xu, Yi Zhang, Chen Zhang, Yasong Du, Wen-Hao Zhou, Xiaoqun Wang, Zilong Qiu
Haploinsufficiency of autism candidate gene NUAK1 impairs cortical development and behavior. Virginie Courchet, Amanda J Roberts, Peggy Del Carmine, Tommy L Lewis Jr., Franck Polleux, Julien Courchet
The dyslexia susceptibility KIAA0319 gene shows a highly specific expression pattern during zebrafish development supporting a role beyond neuronal migration. Monika Gostic, Angela Martinelli, Carl Tucker, Zhengyi Yang, Federico Gasparoli, Kishan Dholakia, Keith Sillar, Javier Tello, Silvia Paracchini
Accelerated brain aging towards transcriptional inversion in a zebrafish model of familial Alzheimer’s disease. Nhi Hin, Morgan Newman, Jan Kaslin, Alon M. Douek, Amanda Lumsden, Xin-Fu Zhou, Noralyn B. Mañucat-Tan, Alastair Ludington, David L. Adelson, Stephen Pederson, Michael Lardelli
Ratiometric assays of autophagic flux in zebrafish for analysis of familial Alzheimer’s disease-like mutations. Haowei Jiang, Morgan Newman, Dhanushika Ratnayake, Michael Lardelli
Increased autophagy in ephrinB2 deficient osteocytes is associated with hypermineralized, brittle bones. Christina Vrahnas, Toby A Dite, Niloufar Ansari, Blessing Crimeen-Irwin, Huynh Nguyen, Mark R Forwood, Yifang Hu, Mika Ikegame, Keith R Bambery, Cyril Petibois, Mark J Tobin, Gordon K Smyth, Jonathan S Oakhill, T John Martin, Natalie A Sims
Evo-devo & evo
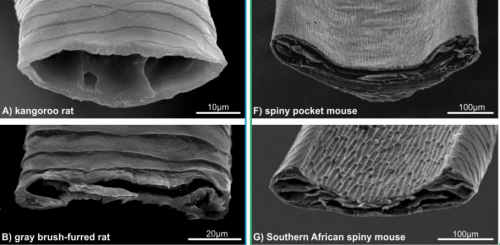
Divergent genetic mechanisms lead to spiny hair in mammals. Gislene L. Goncalves, Renan Maestri, Gilson R.P. Moreira, Marly A.M. Jacobi, Thales R.O. Freitas, Hopi E. Hoekstra
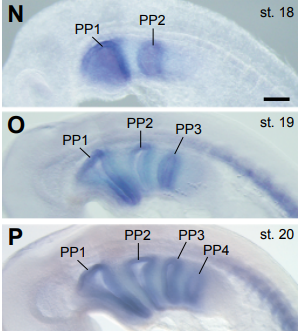
Endoderm Morphogenesis Reveals Integration of Distinct Processes in the Development and Evolution of Pharyngeal Arches. Shinji Takada, Kazunori Okada, Hiroshi Wada
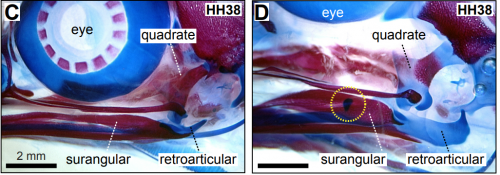
Developmental Mechanisms Linking Form and Function During Jaw Evolution. Katherine C Woronowicz, Stephanie E Gline, Safa T Herfat, Aaron J Fields, Richard A Schneider
An evolutionary-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. Matthias Christian Vogg, Leonardo Beccari, Laura Iglesias Olle, Christine Rampon, Sophie Vriz, Chrystelle Perruchoud, Yvan Wenger, Brigitte Galliot
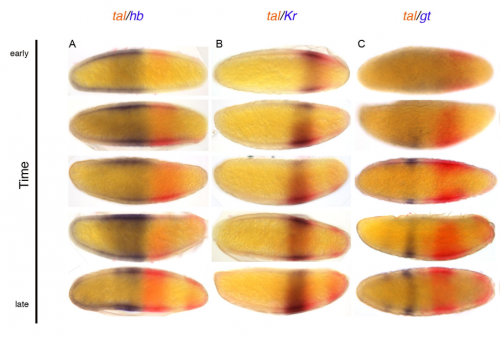
tarsal-less is expressed as a gap gene but has no gap gene phenotype in the moth midge Clogmia albipunctata. eva jimenez-guri, Karl Wotton, Johannes Jaeger
Evidence of adhesion and signaling roles for β-catenin in the sponge Ephydatia muelleri. Klaske J. Schippers, Scott A. Nichols
From spiral cleavage to bilateral symmetry: The developmental cell lineage of the annelid brain. Pavel Vopalensky, Maria Antonietta Tosches, Kaia Achim, Mette Handberg-Thorsager, Detlev Arendt
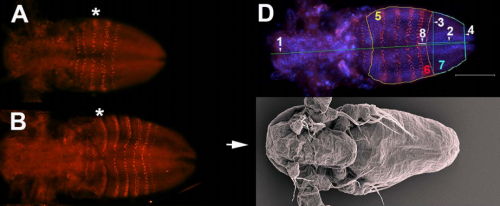
Elongation during segmentation shows axial variability, low mitotic rates, and synchronized cell cycle domains in the crustacean, Thamnocephalus platyurus. Savvas J Constantinou, Nicole Duan, Ariel D. Chipman, Lisa M. Nagy, Terri A. Williams
Changes in the neuropeptide complement correlate with nervous system architectures in xenacoelomorphs. Daniel Thiel, Mirita Franz-Wachtel, Felipe Aguilera, Andreas Hejnol
Intergenerational epigenetic inheritance in reef-building corals. Yi Jin Liew, Emily J. Howells, Xin Wang, Craig T. Michell, John A. Burt, Youssef Idaghdour, Manuel Aranda
Expanding the genetic toolkit of Tribolium castaneum. Johnathan C Rylee, Dylan J Siniard, Kaitlin Doucette, Gabriel E Zentner, Andrew Zelhof
Convergent evolution of gene expression in two high-toothed stickleback populations. James C Hart, Nicholas A Ellis, Michael B Eisen, Craig T Miller
Identification of general patterns of sex-biased expression in Daphnia, a genus with environmental sex determination. Cécile Molinier, Céline Reisser, Peter Fields, Adeline Segard, Yan Galimov, Christoph R. Haag
Genomic imprinting mediates dosage compensation in a young plant XY system. Aline Muyle, Niklaus Zemp, Cecile Fruchard, Radim Cegan, Jan Vrana, Clothilde Deschamps, Raquel Tavares, Franck Picard, Roman Hobza, Alex Widmer, Gabriel Marais
Delegating sex: differential gene expression in stolonizing syllids uncovers the hormonal control of reproduction in Annelida. Patricia Alvarez-Campos, Nathan Kenny, Aida Verdes, Rosa Maria Fernandez, Marta Novo, Gonzalo Giribet, Ana Riesgo
Concurrent duplication of the Cid and Cenp-C genes in the Drosophila subgenus with signatures of subfunctionalization and male germline-biased expression. Jose R. Teixeira, Guilherme B. Dias, Marta Svartman, Alfredo Ruiz, Gustavo C. S. Kuhn
Convergent regulatory evolution and the origin of flightlessness in palaeognathous birds. Timothy B. Sackton, Phil Grayson, Alison Cloutier, Zhirui Hu, Jun S. Liu, Nicole E. Wheeler, Paul P. Gardner, Julia A. Clarke, Allan J. Baker, Michele Clamp, Scott V. Edwards

Genotyping-by-sequencing supports a genetic basis for alpine wing-reduction in a New Zealand stonefly. Andrew J. Veale, Brodie J. Foster, Peter K. Dearden, Jon M. Waters
Transposable elements generate regulatory novelty in a tissue specific fashion. Marco Trizzino, Aurelie Kapusta, Christopher Brown
The effects of haploid selection on Y chromosome evolution in a dioecious plant. George Sandler, Felix E.G. Beaudry, Spencer C.H. Barrett, Stephen I. Wright
Adaptive Reduction of Male Gamete Number in a Selfing Species. Takashi Tsuchimatsu, Hiroyuki Kakui, Misako Yamazaki, Cindy Marona, Hiroki Tsutsui, Afif Hedhly, Dazhe Meng, Yutaka Sato, Thomas Stadler, Ueli Grossniklaus, Masahiro M. Kanaoka, Michael Lenhard, Magnus Nordborg, Kentaro K. Shimizu
Firefly genomes illuminate parallel origins of bioluminescence in beetles. Timothy R Fallon, Sarah E Lower, Ching-Ho Chang, Manabu Bessho-Uehara, Gavin J Martin, Adam J Bewick, Megan Behringer, Humberto J Debat, Isaac Wong, John C Day, Anton Suvorov, Christian J Silva, Kathrin F Stanger-Hall, David W Hall, Robert J. Schmitz, David R Nelson, Sara Lewis, Shuji Shigenobu, Seth M Bybee, Amanda M Larracuente, Yuichi Oba, Jing-Ke Weng

The Aquilegia genome: adaptive radiation and an extraordinarily polymorphic chromosome with a unique history. Daniele Filiault, Evangeline Ballerini, Terezie Mandakova, Gokce Akoz, Nathan Derieg, Jeremy Schmutz, Jerry Jenkins, Jane Grimwood, Shengqiang Shu, Richard Hayes, Uffe Hellsten, Kerrie Barry, Juyiang Yan, Sirma Mihaltcheva, Miroslava Karafiatova, Viktoria Nizhynska, Martin Lysak, Scott Hodges, Magnus Nordborg
The genome of the water strider Gerris buenoi reveals expansions of gene repertoires associated with adaptations to life on the water. David Armisen, Rajendhran Rajakumar, Markus Friedrich, Joshua B Benoit, Hugh M Robertson, Kristen A Panfilio, Seung-Joon Ahn, Monica F Poelchau, Hsu Chao, Huyen Dinh, HarshaVardhan Doddapaneni, Shannon Dugan-Perez, Richard A Gibbs, Daniel ST Hughes, Yi Han, Sandra L Lee, Shwetha C Murali, Donna M Muzny, Jiaxin Qu, Kim C Worley, Monica Munoz-Torres, Ehab Abouheif, Francois Bonneton, Travis Chen, Li-Mei Chiang, Christopher P. Childers, Andrew G Cridge, Antonin JJ Crumiere, Amelie Decaras, Elise M Didion, Elizabeth Duncan, Elena N Elpidina, Marie-Julie Fave, Cedric Finet, Chris GC Jacobs, Alys Jarvela, Emily J Jennings, Jeffery W Jones, Maryna P Lesoway, Mackenzie Lovegrove, Alexander Martynov, Brenda Oppert, Angelica Lilico-Ouachour, Arjuna Rajakumar, Peter N Refki, Andrew J Rosendale, Maria Emilia Santos, William Toubiana, Maurijn van der Zee, Iris M Vargas Jentzsch, Aidamalia Vargas Lowman, Severine Viala, Stephen Richards, Abderrahman Khila
Cell biology
F-actin patches nucleated on chromosomes coordinate capture by microtubules in oocyte meiosis. Mariia Burdyniuk, Andrea Callegari, Masashi Mori, Francois Nedelec, Peter Lenart
Rab-mediated trafficking in the secondary cells of Drosophila male accessory glands and its role in fecundity. Elodie Prince, Marko Brankatschk, Benjamin Kroeger, Clive Wilson, Dragan Gligorov, Suzanne Eaton, Francois Karch, Robert K Maeda
Persistent cell motility requires transcriptional feedback of cytoskeletal – focal adhesion equilibrium by YAP/TAZ. Devon E Mason, James H Dawahare, Trung Dung Nguyen, Yang Lin, Sherry L. Voytik-Harbin, Pinar Zorlutuna, Mervin E Yoder, Joel D Boerckel
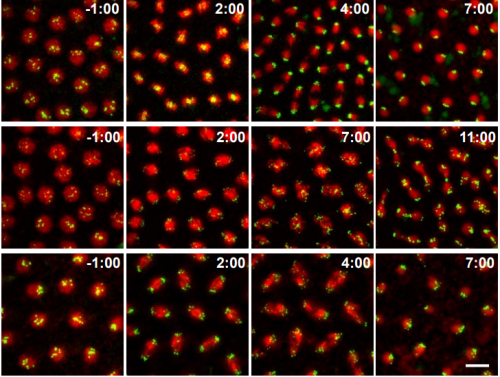
Absence of the Spindle Assembly Checkpoint restores mitotic fidelity upon loss of sister chromatid cohesion. Rui D Silva, Mihailo Mirkovic, Leonardo G Guilgur, Om S. Rathore, Rui Gonçalo Martinho, Raquel A Oliveira
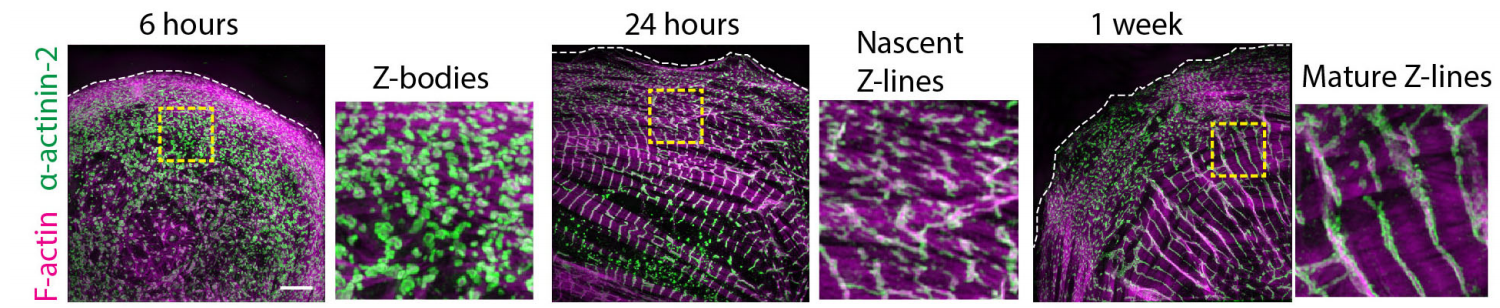
Focal adhesion kinase regulates early steps of myofibrillogenesis in cardiomyocytes. Nilay Taneja, Abigail C Neininger, Matthew R Bersi, W David Merryman, Dylan T Burnette
Recruitment of Two Dyneins to an mRNA-Dependent Bicaudal D Transport Complex. Thomas E. Sladewski, Neil Billington, M. Yusuf Ali, Carol S. Bookwalter, Hailong Lu, Elena B. Krementsova, Trina A. Schroer, Kathleen M. Trybus
RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. Mark A McClintock, Carly I Dix, Christopher M Johnson, Stephen H McLaughlin, Rory J Maizels, Ha Thi Hoang, Simon L Bullock
Dynein-2 intermediate chains play crucial but distinct roles in primary cilia formation and function. Laura Vuolo, Nicola L Stevenson, Kate Heesom, David J Stephens
Regulator of calcineurin-2 is a centriolar protein with a role in cilia length control. Nicola Stevenson, Dylan Bergen, Amadeus Xu, Emily Wyatt, Freya Henry, Janine McCaughey, Laura Vuolo, Chrissy Hammond, David Stephens
Actin and Src-family kinases regulate nuclear YAP1 and its export. Nil Ege, Anna M Dowbaj, Ming Jiang, Michael Howell, Robert P Jenkins, Erik Sahai
Rotational 3D mechanogenomic Turing patterns of human colon Caco-2 cells during differentiation. Gen Zheng, Alexandr A. Kalinin, Ivo D. Dinov, Walter Meixner, Shengtao Zhu, John W. Wile
Collagen stiffness modulates MDA-MB231 cell metabolism through adhesion-mediated contractility. Emma J. Mah, Gabrielle E. McGahey, Albert F. Yee, Michelle A. Digman
Tunable molecular tension sensors reveal extension-based control of vinculin loading. Andrew S LaCroix, Andrew D Lynch, Matthew E Berginski, Brenton D Hoffman
CLIC4 is regulated by RhoA-mDia2 signaling through Profilin-1 binding to modulate filopodia length. Elisabetta Argenzio, Katarzyna M Kedziora, Leila Nahidiazar, Tadamoto Isogai, Anastassis Perrakis, Kees Jalink, Wouter H Moolenaar, Metello Innocenti
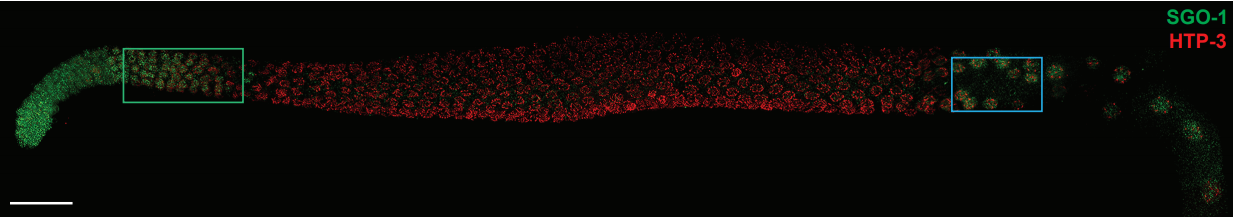
Shugoshin is essential for meiotic prophase checkpoints in C. elegans. Tisha Bohr, Christian R. Nelson, Stefani Giacopazzi, Piero Lamelza, Needhi Bhalla
Shugoshin protects centromere pairing and promotes segregation of non-exchange partner chromosomes in meiosis. Luciana Previato, Emily L Kurdzo, Craig A Eyster, Mara N Gladstone, Hoa H Chuong, Elena Llano, Alberto M Pendas, Roberto J Pezza, Dean S Dawson
A positive feedback-based mechanism for constriction rate acceleration during cytokinesis in C. elegans. Renat Khaliullin, Rebecca Green, Linda Shi, Michael Berns, J. Sebastian Gomez-Cavazos, Arshad Desai, Karen Oegema
Cell-matrix adhesion controls Golgi organization and function by regulating Arf1 activation in anchorage-dependent cells. Vibha Singh, Chaitanya Erady, Nagaraj Balasubramanian
Modelling
Spatial dynamics of flower organ formation. Yuriria Cortes Poza, Pablo Padilla Longoria, Elena Alvarez Buylla
Mathematical modelling of cortical neurogenesis reveals that the founder population does not necessarily scale with neuronal output. Noemi Picco, Fernando Garcia-Moreno, Thomas Woolley, Philip Maini, Zoltan Molnar
From jamming to collective cell migration through a boundary induced transition. Oleksandr Chepizhko, Maria Chiara Lionetti, Chiara Malinverno, Giorgio Scita, Stefano Zapperi, Caterina A. M. La Porta
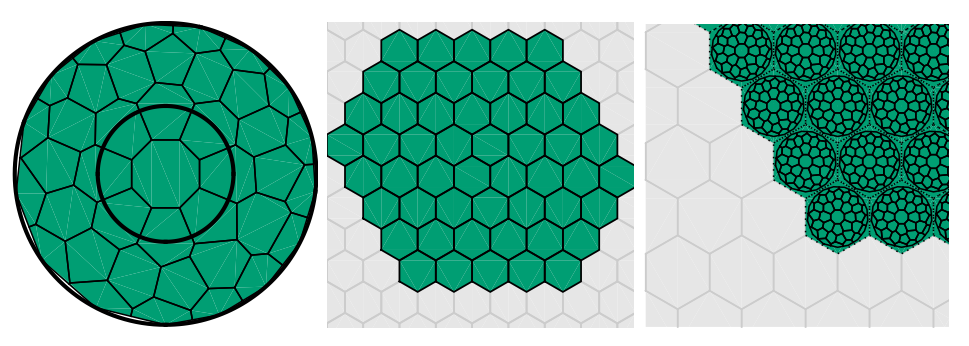
Stochastic simulation of pattern formation in growing tissue: a multilevel approach. Stefan Engblom
Self-organization principles of intracellular pattern formation. Jacob Halatek, Fridtjof Brauns, Erwin Frey
Modeling cell-substrate de-adhesion dynamics under fluid shear. Renu Maan, Gautam I Menon, Pramod A Pullarkat
Theory of Morphogenesis. Andrey Minarsky, Nadya Morozova, Robert Penner, Christophe Soule
A mathematical model of the metabolism of a cell. Self-organization and chaos. V.I. Grytsay, I.V. Musatenko
Transition State Characteristics During Cell Differentiation. Rowan Brackston, Eszter Lakatos, Michael P.H. Stumpf
Riding the Cell Jamming Boundary: Geometry, Topology, and Phase of Human Corneal Endothelium. Nigel H Brookes
Stochasticity of cellular growth: sources, propagation and consequences. Philipp Thomas, Guillaume Terradot, Vincent Danos, Andrea Weisse
Statistical Inference in Cell Lineage Trees. Damien G Hicks, Terence P Speed, Mohammed Yassin, Sarah M Russell
Generative adversarial networks uncover epidermal regulators and predict single cell perturbations. Arsham Ghahramani, Fiona M Watt, Nicholas M Luscombe
Information Isometry Technique Reveals Organizational Features in Developmental Cell Lineages. Bradly John Alicea, Thomas E. Portegys, Richard Gordon
In silico mechanobiochemical modeling of morphogenesis in cell monolayers. Bahador Marzban, Xiao Ma, Xiaoliang Qing, Hongyan Yuan
Noise-driven Cell Differentiation and the Emergence of Organization. Hadiseh Safdari, Ata Kalirad, Cristian Picioreanu, Rouzbeh Tusserkani, Bahram Goliaei, Mehdi Sadeghi
Tools & resources
| Imaging etc.
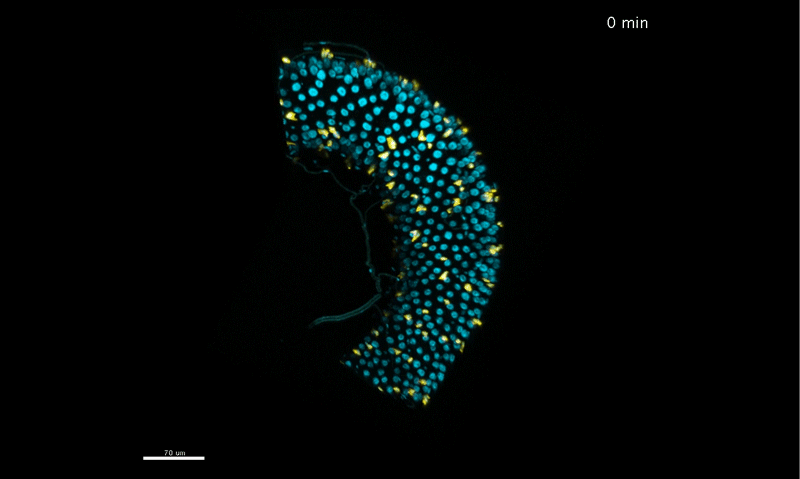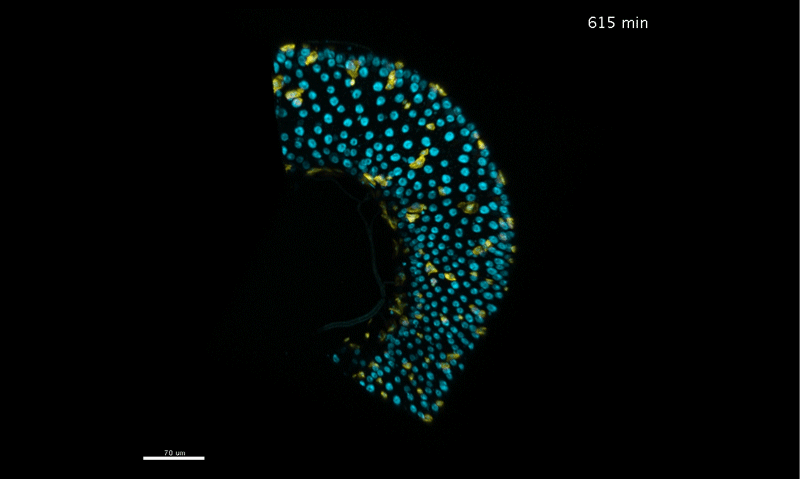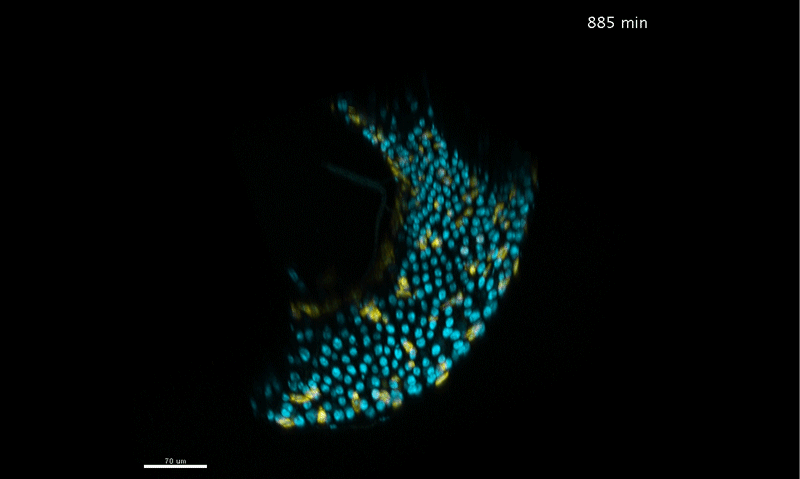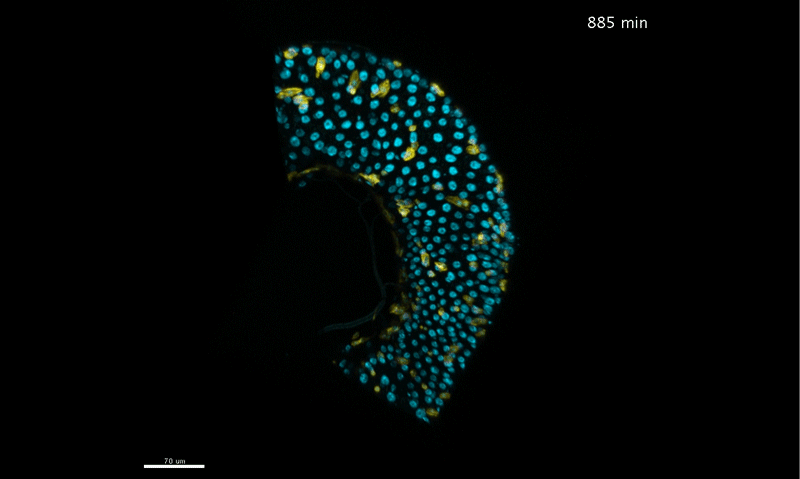
Long-term live imaging of the Drosophila adult midgut reveals real-time dynamics of cell division, differentiation, and loss. Judy Martin, Erin Nicole Sanders, Paola Moreno-Roman, Shruthi Balachandra, XinXin Du, Leslie Ann Jaramillo Koyama, Lucy Erin O’Brien
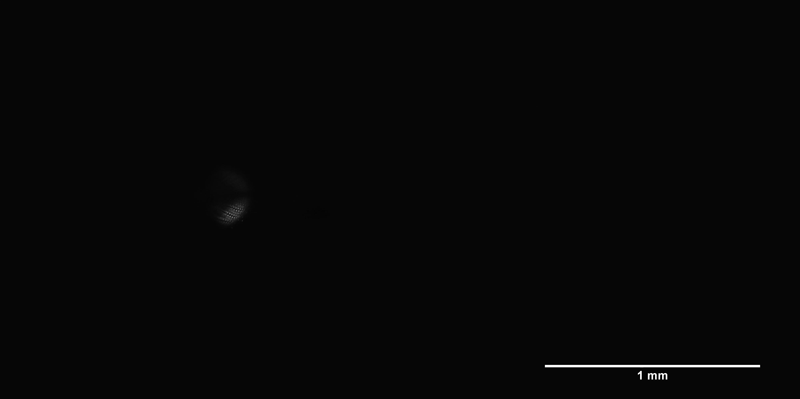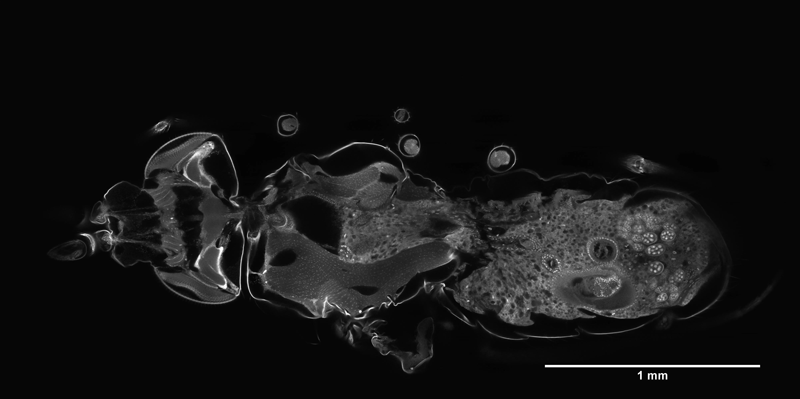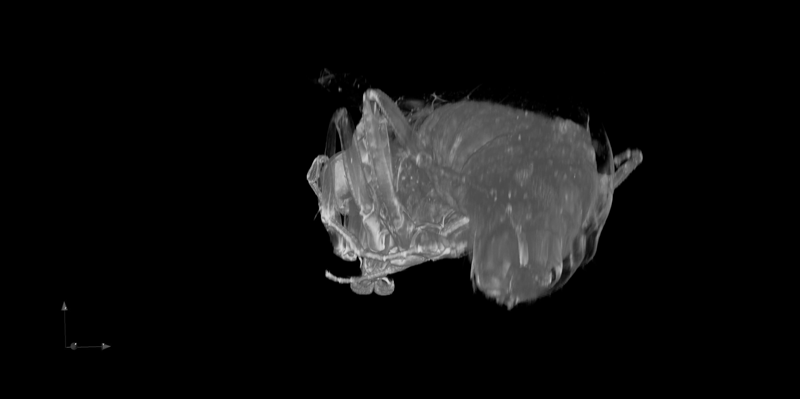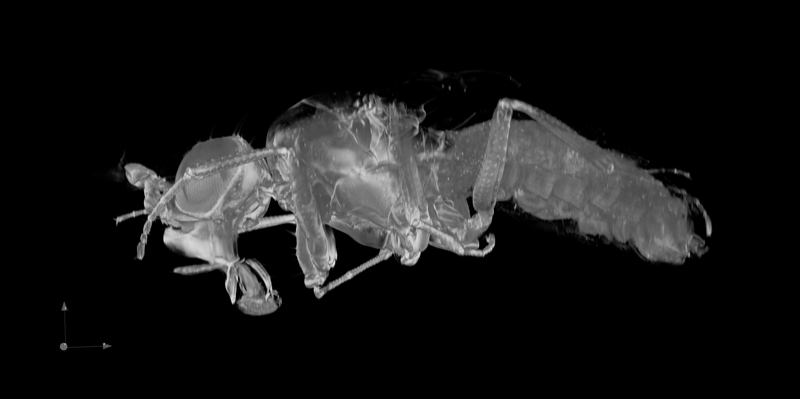
Application of the Mesolens for sub-cellular resolution imaging of intact larval and whole adult Drosophila. Gail McConnell, William B. Amos
Brain Development: Machine Learning Analysis Of Individual Stem Cells In Live 3D Tissue. Martin Hailstone, Lu Yang, Dominic Waithe, Tamsin J. Samuels, Yoav Arava, Tomasz Dobrzycki, Richard M. Parton, Ilan Davis
Multidirectional digital scanned light-sheet microscopy enables uniform fluorescence excitation and contrast-enhanced imaging. Adam K Glaser, Ye Chen, Chengbo Yin, Linpeng Wei, Lindsey Barner, Nicholas Reder, Jonathan Liu
Forget Pixels: Adaptive Particle Representation of Fluorescence Microscopy Images. Bevan L. Cheeseman, Ulrik Günther, Mateusz Susik, Krzysztof Gonciarz, Ivo F. Sbalzarini
Image-based methods for phenotyping growth dynamics and fitness in Arabidopsis thaliana. Francois Vasseur, George Wang, Justine Bresson, Rebecca Schwab, Detlef Weigel
Mid-infrared optoacoustic microscopy with label-free chemical contrast in living cells and tissues. Miguel A. Pleitez, Asrar Ali Khan, Josefine Reber, Andriy Chmyrov, Markus Seeger, Marcel Scheideler, Stephan Herzig, Vasilis Ntziachristos
High Numerical Aperture Epi-illumination Selective Plane Illumination Microscopy. Bin Yang, Yina Wang, Siyu Feng, Veronica Pessino, Nico Stuurman, Bo Huang
Imaging the Developing Heart: Synchronized Timelapse Microscopy During Developmental Changes. Carl J. Nelson, Charlotte Buckley, John J. Mullins, Martin A. Denvir, Jonathan Taylor
4D blood flow mapping using SPIM-microPIV in the developing zebrafish heart. Vytautas Zickus, Jonathan M. Taylor
Quantitative mapping of fluorescently tagged cellular proteins using FCS-calibrated four dimensional imaging. Antonio Z Politi, Yin Cai, Nike Walther, M. Julius Hossain, Birgit Koch, Malte Wachsmuth, Jan Ellenberg
ChromoTrace: Reconstruction of 3D Chromosome Configurations by Super-Resolution Microscopy. Carl Barton, Sandro Morganella, Oeyvind Oedegaard, Stephanie Alexander, Jonas Ries, Tomas Fitzgerald, Jan Ellenberg, Ewan Birney
Estimating numbers of fluorescent molecules in single cells by analysing fluctuations in photobleaching. Elco Bakker, Peter S. Swain
Using Deep Learning for Segmentation and Counting within Microscopy Data. Carlos X. Hernández, Mohammad M. Sultan, Vijay S. Pande
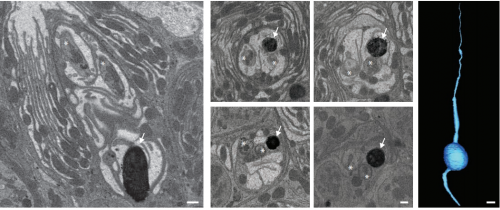
High-quality ultrastructural preservation using cryofixation for 3D electron microscopy of genetically labeled tissues. Tin Ki Tsang, Eric A Bushong, Daniela Boassa, Junru Hu, Benedetto Romoli, Sebastien Phan, Davide Dulcis, Chih-Ying Su, Mark H Ellisman
| Genome tools
In vivo CRISPR-Cas gene editing with no detectable genome-wide off-target mutations. Pinar Akcakaya, Maggie L. Bobbin, Jimmy A. Guo, Jose Malagon Lopez, M. Kendell Clement, Sara P. Garcia, Mick D. Fellows, Michelle J. Porritt, Mike A. Firth, Alba Carreras, Tania Baccega, Frank Seeliger, Mikael Bjursell, Shengdar Q. Tsai, Nhu T. Nguyen, Roberto Nitsch, Lorenz Mayr, Luca Pinello, Mohammad Bohlooly-Y, Martin J. Aryee, Marcello Maresca, J. Keith Joung
No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. Vivek Iyer, Katharina Boroviak, Mark Thomas, Brendan Doe, Edward Ryder, David Adams
On the design of CRISPR-based single cell molecular screens. Andrew J Hill, José L McFaline-Figueroa, Lea M Starita, Molly J Gasperini, Kenneth A Matreyek, Jonathan Packer, Dana Jackson, Jay Shendure, Cole Trapnell
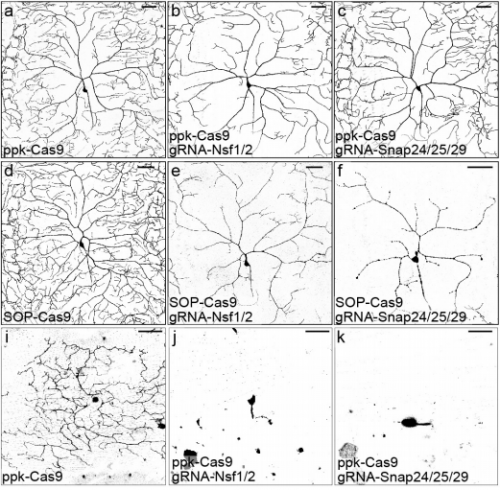
Highly efficient CRISPR/Cas9-mediated tissue specific mutagenesis in Drosophila. Amy R Poe, Bei Wang, Maria L Sapar, Hui Ji, Kailyn Li, Tireniolu Onabajo, Rushaniya Fazliyeva, Mary Gibbs, Yue Qiu, Yuzhao Hu, Chun Han
Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence repair outcome. Eirini M Kallimasioti-Pazi, Keerthi Thelakkad Chathoth, Gillian C Taylor, Alison Meynert, Tracy Ballinger, Martijn J Kelder, Sebastien Lalevee, Ildem Sanli, Robert Feil, Andrew J Wood
Machine learning of stem cell identities from single-cell expression data via regulatory network archetypes. Patrick S. Stumpf, Ben D. MacArthur
Rapid low-cost assembly of the Drosophila melanogaster reference genome using low-coverage, long-read sequencing. Edwin A. Solares, Mahul Chakraborty, Danny E. Miller, Shannon Kalsow, Kate E. Hall, Anoja G. Perera, J.J. Emerson, R. Scott Hawley
High-quality genome assemblies of 15 Drosophila species generated using Nanopore sequencing. Danny E. Miller, Cynthia Staber, Julia Zeitlinger, R. Scott Hawley
Multiple laboratory mouse reference genomes define strain specific haplotypes and novel functional loci. Jingtao Lilue, Anthony G Doran, Ian T Fiddes, Monica Abrudan, Joel Armstrong, Ruth Bennett, William Chow, Joanna Collins, Anne Czechanski, Petr Danecek, Mark Diekhans, Dirk-Dominic Dolle, Matt Dunn, Richard Durbin, Dent Earl, Anne Ferguson-Smith, Paul Flicek, Jonathan Flint, Adam Frankish, Beiyuan Fu, Mark Gerstein, James Gilbert, Leo Goodstadt, Jennifer Harrow, Kerstin Howe, Mikhail Kolmogorov, Stefanie Koenig, Chris Lelliott, Jane Loveland, Richard Mott, Paul Muir, Fabio Navarro, Duncan Odom, Naomi Park, Sarah Pelan, Son K Phan, Michael Quail, Laura Reinholdt, Lars Romoth, Lesley Shirley, Cristina Sisu, Marcela Sjoberg-Herrera, Mario Stanke, Charles Steward, Mark Thomas, Glen Threadgold, David Thybert, James Torrance, Kim Wong, Jonathan Wood, Fengtang Yang, David J Adams, Benedict Paten, Thomas M Keane
Programmable single and multiplex base-editing in Bombyx mori using RNA-guided cytidine deaminases. Qingyou Xia, Yufeng Li, Sanyuan Ma, Le Sun, Tong Zhang, Jiasong Chang, Wei Lu, Xiaoxu Chen, Yue Liu, Xiaogang Wang, Run Shi, Ping Zhao
A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Christian Pflueger, Dennis Tan, Tessa Swain, Trung Viet Nguyen, Jahnvi Pflueger, Christian Nefzger, Jose M Polo, Ethan Ford, Ryan Lister
Addressing the looming identity crisis in single cell RNA-seq. Megan Crow, Anirban Paul, Sara Ballouz, Z. Josh Huang, Jesse Gillis
Enhancing resolution of natural methylome reprogramming behavior in plants. Robersy Sanchez, Xiaodong Yang, Hardik Kundariya, Jose Raul Barreras, Yashitola Wamboldt, Sally Mackenzie
Verified the effectiveness of AsCpf1 system in a variety of vertebrate species. Zhuo Li, Zhaoying Shi, Nana Fan, Yongqiang Chen, Jing Guo, Jingchun Wu, Hong Song, Shilong Chu, Kunlun Mo, Bentian Zhao, Zhen Ouyang, Dandan Tian, Shaoyang Zhao, Jieying Zhu, Jiekai Chen, Yonglong Chen, Liangxue Lai, Duanqing Pei
CRISPR-DT: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Houxiang Zhu, Chun Liang
CONSTRUCTION OF WHOLE GENOMES FROM SCAFFOLDS USING SINGLE CELL STRAND-SEQ DATA. Mark Hills, Ester Falconer, Kieran O’Neil, Ashley Sanders, Kerstin Howe, Victor Guryev, Peter M Lansdorp
Cas-CLIP: a method for customizing pooled CRISPR libraries. Jiyeon Kweon, Da-eun Kim, An-Hee Jang, Yongsub Kim
TOGGLe, a flexible framework for easily building complex workflows and performing robust large-scale NGS analyses. Christine Tranchant-Dubreuil, Sebastien Ravel, Cecile Monat, Gautier Sarah, Abdoulaye Diallo, Laura Helou, Alexis Dereeper, Ndomassi Tando, Julie Orjuela-Bouniol, Francois Sabot
An Interpretable Framework for Clustering Single-Cell RNA-Seq Datasets. Jesse M Zhang, Jue Fan, H. Christina Fan, David Rosenfeld, David N. Tse
Research practice & education
Gender balance in time-keeping at life science conferences. Petra Edlund, Erin M Tranfield, Vera van Noort, Karen Siu Ting, Sofia Tapani, Johanna Hoog
Building a local community of practice in scientific programming for Life Scientists. Sarah L.R. Stevens, Mateusz Kuzak, Carlos Martinez, Aurelia Moser, Petra M. Bleeker, Marc Galland
The impact on authors and editors of introducing Data Availability Statements at Nature journals. Rebecca Grant, Iain Hrynaszkiewicz
Intellectual Synthesis in Mentorship Determines Success in Academic Careers. Jean F Lienard, Titipat Achakulwisut, Daniel E Acuna, Stephen V David
Reproducible big data science: A case study in continuous FAIRness. Ravi K Madduri, Kyle Chard, Mike D’Arcy, Segun C Jung, Alexis Rodriguez, Dinanath Sulakhe, Eric W Deutsch, Cory Funk, Ben Heavner, Matthew Richards, Paul Shannon, Ivo Dinov, Gustavo Glusman, Nathan Price, John D Van Horn, Carl Kesselman, Arthur W Toga, Ian Foster
Twenty steps towards an adequate inferential interpretation of p-values. Norbert Hirschauer, Sven Grüner, Oliver Mußhoff, Claudia Becker
Sci-Hub provides access to nearly all scholarly literature. Daniel S Himmelstein, Ariel R Romero, Jacob G Levernier, Thomas A Munro, Stephen R McLaughlin, Bastian Greshake Tzovaras, Casey S Greene
Sharing is not erring: Pseudo-reciprocity in collective search. Imen Bouhlel, eCharley M. Wu, Nobuyuki Hanaki, Robert L. Goldstone
Why not…
FGF21, a liver hormone that inhibits alcohol intake in mice, increases in human circulation after acute alcohol ingestion and sustained binge drinking at Oktoberfest. Susanna Soeberg, Emilie S. Andersen, Niels B. Dalgaard, Ida Jarlhelt, Nina L. Hansen, Nina Hoffmann, Tina Vilsboell, Anne Chenchar, Michal Jensen, Trisha J. Grevengoed, Sam A.J. Trammell, Filip K. Knop, Matthew P. Gillum
Biomechanics of the peafowl’s crest: a potential mechanosensory role for feathers during social display. Suzanne Amador Kane, Daniel Van Beveren, Roslyn Dakin



 (No Ratings Yet)
(No Ratings Yet)
 (3 votes)
(3 votes)