Posted by the Node, on 16 May 2017
Here are the highlights from the current issue of Development:
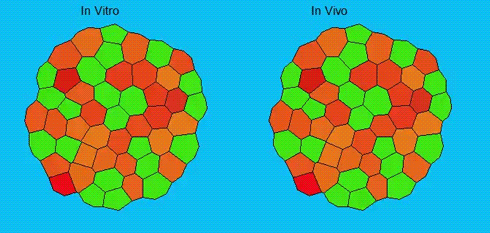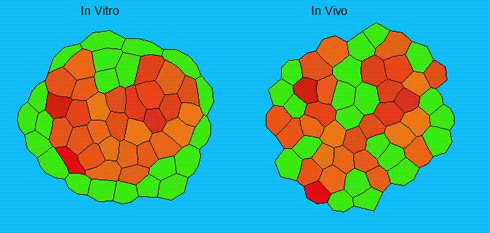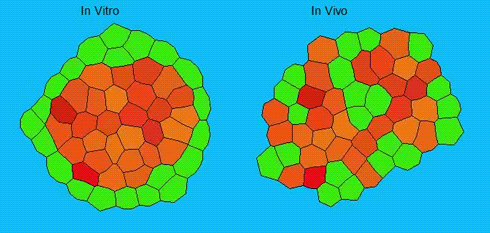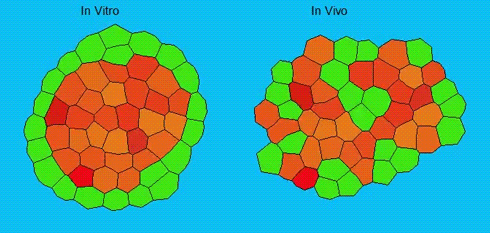
Lineage segregation during gastrulation has long been thought to be driven by differential cell adhesion and cortical tension among cells, which would together lead to a differential tissue surface tension (TST) and the spatial segregation of specific cell types. However, this long-standing hypothesis is mainly based on in vitro work, and it is as yet unclear whether it holds true in vivo. Now, on p. 1798 Carl-Philipp Heisenberg and colleagues assess the role of differential TST in lineage segregation and find that, contrary to in vitro work, differential TST is insufficient to explain progenitor cell segregation and germ layer formation within the in vivo gastrulating zebrafish embryo. In the study, the authors describe their unique version of video force microscopy called 3D CellFit, which allows them to analyse surface tensions in 3D within a living organism. Using this method, the authors show that ectoderm and mesoderm tissues do not, in fact, exhibit differential TST in the gastrula. They further present evidence that the apparent discrepancy between the in vitro and in vivo results is due to a difference in osmolarity between the culture medium and the interstitial fluid that surround the cells. Finally, by inhibiting the function of the small GTPase Rac, a key regulator of protrusion-driven cell migration, the authors show that directed cell migration, rather than differential TST, provides the major mechanism that determines the segregation of the germ layer progenitors.
A crucial phase in neuronal development is the integration of newborn neurons into circuits. The right balance must be struck between excitatory and inhibitory neurons; however, the mechanisms that control inhibitory neuron integration and drive the maturation of inhibitory connectivity remain largely uncharacterized. 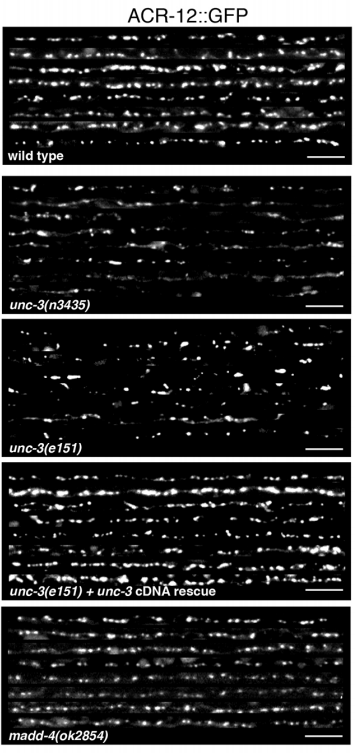 In this issue (p. 1807) Michael Francis and colleagues identify a novel, non-cell-autonomous mechanism that regulates inhibitory neuron synapse formation at the neuromuscular junction (NMJ). The authors examine the electrophysiology and structural organization of GABAergic synapses at the NMJ in a number of different C. elegans mutants with developmental or functional defects in excitatory motor neurons. These analyses reveal that the activity of excitatory cholinergic motor neurons, during a period that coincides with the development of postembryonic GABAergic motor neurons, critically affects the size and distribution of GABAergic pre- and post-synaptic specializations. Furthermore, a severe reduction of cholinergic inputs to newly born GABAergic neurons reduces their synaptic density but increases the synapse size. This study makes an important contribution to our understanding of how neuronal activity impacts synapse development and highlights the functional relationship between excitatory and inhibitory neurons during circuit formation.
In this issue (p. 1807) Michael Francis and colleagues identify a novel, non-cell-autonomous mechanism that regulates inhibitory neuron synapse formation at the neuromuscular junction (NMJ). The authors examine the electrophysiology and structural organization of GABAergic synapses at the NMJ in a number of different C. elegans mutants with developmental or functional defects in excitatory motor neurons. These analyses reveal that the activity of excitatory cholinergic motor neurons, during a period that coincides with the development of postembryonic GABAergic motor neurons, critically affects the size and distribution of GABAergic pre- and post-synaptic specializations. Furthermore, a severe reduction of cholinergic inputs to newly born GABAergic neurons reduces their synaptic density but increases the synapse size. This study makes an important contribution to our understanding of how neuronal activity impacts synapse development and highlights the functional relationship between excitatory and inhibitory neurons during circuit formation.
The directed differentiation of human induced pluripotent stem cells (iPSCs) into mature hepatocytes is a major goal of liver research. The approach relies on the recapitulation of developmental processes, and thus a better understanding of what regulates hepatocyte differentiation is essential in order to produce these cells more efficiently and to a greater maturity. In this issue (p. 1764) Stephen Duncan and colleagues identify heat shock protein 90 beta (HSP90β) as a novel regulator of endoderm-to-hepatocyte conversion in differentiating human iPSC cultures. The authors begin the study by conducting a screen for small molecules that modify the activity of master hepatocyte transcription factor HNF4A, identifying 132 candidate ‘hits’. They then focus on the role of molecular chaperone HSP90β and show how it acts at the post-translational level to stabilize HNF4A, thus controlling its half-life and availability. Targeted CRISPR-CAS9 mutations in the gene encoding HSP90 perturbs HSP90β levels, resulting in a dramatic reduction of HNF4A protein levels and reduced expression of HNF4A target genes. Moreover, these experiments reveal that HSP90β is specifically required for endoderm-to-hepatocyte conversion, and not for endoderm commitment generally. This study uncovers a new player in hepatocyte differentiation, and further highlights the utility of an iPSC differentiation platform coupled with chemical screens to uncover novel developmental mechanisms.
In this issue (p. 1764) Stephen Duncan and colleagues identify heat shock protein 90 beta (HSP90β) as a novel regulator of endoderm-to-hepatocyte conversion in differentiating human iPSC cultures. The authors begin the study by conducting a screen for small molecules that modify the activity of master hepatocyte transcription factor HNF4A, identifying 132 candidate ‘hits’. They then focus on the role of molecular chaperone HSP90β and show how it acts at the post-translational level to stabilize HNF4A, thus controlling its half-life and availability. Targeted CRISPR-CAS9 mutations in the gene encoding HSP90 perturbs HSP90β levels, resulting in a dramatic reduction of HNF4A protein levels and reduced expression of HNF4A target genes. Moreover, these experiments reveal that HSP90β is specifically required for endoderm-to-hepatocyte conversion, and not for endoderm commitment generally. This study uncovers a new player in hepatocyte differentiation, and further highlights the utility of an iPSC differentiation platform coupled with chemical screens to uncover novel developmental mechanisms.
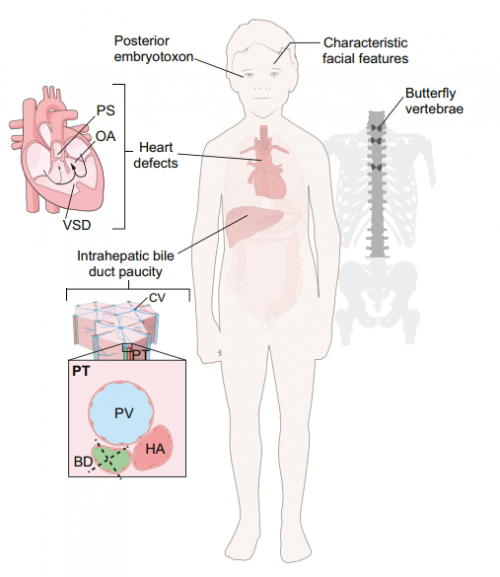
This Review discusses the developmental processes underlying Notch-related congenital disorders in humans, drawing on data from model organisms and genome-sequencing projects, on p. 1743.

In our latest interview, Eric Wieschaus tells us about his Nobel Prize-winning fly screens, his interest in the cell biology of development and his love of painting, on p. 1740.

A retrospective on the life and work of the pioneering Japanese developmental biologist Tokindo Okada, whose research focussed on cell plasticity and transdifferentiation, on p. 1737.
Posted by semrau, on 16 May 2017
Closing Date: 15 March 2021
The Semrau lab
Our lab is interested in the fundamental molecular mechanisms underlying lineage decision-making in stem cells and in vivo. We are fascinated by the question how defined and stable cell types are generated by the interplay of signaling inputs and gene regulatory networks. We study this question by precise quantification of the states of single cells in combination with bioinformatics analysis and machine learning. Based on this quantitative understanding we want to develop new ways to manipulate lineage decisions during in vitro differentiation in precisely controlled ways. Our group is highly interdisciplinary and works at the interface of biology, biophysics, bioinformatics and biomedical sciences.
References:
Semrau, S., van Oudenaarden, A., 2015. Studying Lineage Decision-Making In Vitro: Emerging Concepts and Novel Tools. Annu. Rev. Cell Dev. Biol. 31, 317–345. doi:10.1146/annurev-cellbio-100814-125300
Semrau, S., Goldmann, J., Soumillon, M., Mikkelsen, T.S., Jaenisch, R., van Oudenaarden, A., 2016. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. bioRxiv 068288. doi:10.1101/068288
Semrau, S., Crosetto, N., Bienko, M., Boni, M., Bernasconi, P., Chiarle, R., van Oudenaarden, A., 2014. FuseFISH: Robust Detection of Transcribed Gene Fusions in Single Cells. Cell Reports 6, 18–23. doi:10.1016/j.celrep.2013.12.002
Project and key responsibilities
The available postdoc project aims to create a single-cell atlas of the human embryonic kidney. Information about the transcriptional profiles and locations of all cell types in the embryonic kidney will improve our understanding of kidney development and will provide an important benchmark for kidney organoids. In this project you will be responsible for performing single-cell RNA-seq and single-molecule FISH measurements of human embryonic kidney samples. The necessary experimental techniques are established in our lab and samples will be provided by our collaborators. In particular, you will dissociate the tissue and prepare single-cell RNA-seq libraries with the drop-seq technique (Macosko et al., Cell, 2015). You will analyze the RNA-seq data (potentially together with a bioinformatics collaborator) and identify cell types using state-of-the-art machine learning tools. Based on these results you will define a set of marker genes that will allow you to locate cell types by single-molecule FISH in intact tissue sections. This comprehensive spatial molecular data set will then allow you, for example, to establish intercellular signaling networks.
Selection criteria
Research at our department
Our lab is part of the Leiden Institute of Physics (http://www.physics.leidenuniv.nl) and situated at the Leiden Cell Observatory (http://cellobservatory.leidenuniv.nl). The Cell Observatory is a highly collaborative community dedicated to the visualization and understanding of the fundamental molecular mechanisms of life, which is part of the core scientific profile of Leiden University. The Cell Observatory houses state-of-the-art bio-imaging facilities shared among the member labs, which actively develop new methods for the quantitative measurement of single-cell properties.
Information
More information about our lab can be found at http://www.semraulab.com/.
Enquiries can be made to Dr. Stefan Semrau (semrau@physics.leidenuniv.nl).
Information about the Faculty of Science can be found at http://www.science.leidenuniv.nl/index.php/english/ and about Leiden University at http://workingat.leiden.edu/.
Applications
To apply for this vacancy, please send an email to Dr. Stefan Semrau (semrau@physics.leidenuniv.nl) until June 18. Please include your curriculum vitae, a letter of motivation and the names of 3 potential references.
Posted by Chong-Morrison, on 16 May 2017
“It finally got accepted!”, followed by “It’s finally out!” about a month later. I am certain this ‘finally’ feeling about their paper is very familiar to those well-acquainted with the peer review process, and it was no different for our recently published Resource article. The ‘biotagging paper’, as we call it within the Sauka-Spengler lab, is the culmination of several years’ of hard (and often frustrating) work that eventually paid off in more (unexpected) ways than one. Tatjana spearheaded the initial work for biotagging while still at Caltech, by transferring components and approaches she developed in the chicken system into the zebrafish. She worked together with Le and Tatiana, then postdoctoral fellows at Caltech, before the rest of us joined in for the lengthy optimisation, submission and review stage.
Biotagging is essentially an encompassing term for our Do-It-Yourself (DIY) in vivo biotinylation system for zebrafish researchers, which can be utilised in creative ways to suit specific biological needs. In vivo biotinylation was first employed in mouse (de Boer et al., 2003) by John Strouboulis when he was in Frank Grosveld’s lab and then applied for use in nuclei isolation from Arabidopsis thaliana by Roger Deal and Steven Henikoff (Deal and Henikoff, 2010). The technique was also applied to the nematode worm at around the same time (Ooi et al. 2009). The core of the technique lies in the ability of bacterial biotin ligase (BirA) to biotinylate an Avi-tagged protein-of-interest. In our binary biotagging system, the researcher decides where BirA will be expressed, which protein is Avi-tagged, and then generates transgenic lines that express these components. Crossing BirA-driver and Avi-effector heterozygous lines will give rise to ~25% of double-alleled offspring, where biotinylation of the Avi-tagged protein product only occurs in cells that also express BirA. The sky is the limit when it comes to the combinations of BirA/Avi that one can use. In the paper, we present a ‘starter’ toolkit consisting of multiple tissue-(neural crest, heart, blood) and cellular compartment-specific (ribosomes, nuclei) transgenic lines, as well as constructs to make your own lines.
The deconstruction (and reconstruction) of biotagging
The elegance of in vivo biotinylation means that we are not the only group to perform this method in vertebrates. For example, Michael Housley from Stainier lab (Housley et al. 2014) utilised in vivo biotinylation in zebrafish to apply the TRAP (Translating Ribosome Affinity Purification) method developed by Myriam Heiman and colleagues (Heiman et al., 2008). In vivo biotinylation experiments are not ‘difficult’ per se, but we found that obtaining a clear difference between nuclear and polyribosomal data required a remarkable amount of troubleshooting and optimisation. Our patience paid off, as this was rewarded by a wealth of information provided by a high resolution view into the migratory neural crest nascent (nuclear) and polyribosomal transcriptomes from ~200k cells.
In fact, the entire optimisation process came about by accident. In the paper, we described our surprising results when comparing the nuclear transcriptome of Sox10-positive cells at 16-18ss (migratory neural crest) to a ubiquitous control. By looking at both non-poly and polyadenylated transcripts (whole nuclear transcriptomes), our data did not yield any statistically significant neural crest-specific signature, which is what one would expect, as the enriched transcripts should be neural crest-specific. On the other hand, analysis of polyadenylated nuclear transcripts at 24hpf yielded a neural crest-specific signature. This led to further pain-staking deconstruction of our technique where, months later, we eventually came to the surprisingly simple but crucial element for the protocol to be as consistent as it is today – ensuring the complete lysis of cells (by using hypotonic buffer in excess) to release subcellular compartments into the lysate and minimise the presence of intact cell surface membranes. It is also worth noting, that a key element to the success of our protocol was the usage of an Avi-tagged chicken nuclear envelope protein, RanGAP, to label nuclei. Weirdly enough, chicken RanGAP expressed in zebrafish localised to the nuclei, but zebrafish RanGAP did not.
Having reconstructed the method, we were now eager to repeat the previous 16-18ss neural crest experiment. Imagine our initial dismay when the results were…strikingly similar. However, this was soon replaced by curiosity that drove us to carefully re-examine our results and try to figure out what IS actually going on…
Biotagging of migratory neural crest nuclei transcriptome reveals…what?
The brainstorming sessions were remarkably memorable. They were always long, often ‘lively’ as we picked at each other’s brains, and at times quite outrageous as frustrations ran high. It didn’t take us very long to notice that bidirectional transcription at non-coding regions was enriched in neural crest nuclei. However, it was a long journey after that, as we tried to quantify the phenomenon genome-wide, reproduce what we saw, believe in what we saw, and build our findings into a coherent story. Ultimately, we needed to drive home our main message – that bidirectional transcription at non-coding regions is tissue-specific, thus introducing a new method to detect active regulatory elements. These elements form the molecular signature of neural crest cells, which is traditionally based on the expression of protein-coding genes that are mainly transcription factors. We were also excited to find developmentally regulated long non-coding RNAs and transposable elements.
In short, we are proud of what we have managed to achieve with biotagging. The journey may have been long and arduous, but we have learned a lot from this project. We hope that we have provided a cool new system that includes a fully optimised tool (plasmids on Addgene) with clean protocols (available on the Resources page of our lab website), handy transgenic lines to get started with, as well as analysis pipelines tailored to biotagging datasets. Having worked out the technical intricacies of this system, this toolkit allows the zebrafish community (including us!) to study specific cellular populations in vivo on the systems level, tackling biological questions that could be important to development and disease.
Le A. Trinh, Vanessa Chong-Morrison, Daria Gavriouchkina, Tatiana Hochgreb-Hägele, Upeka Senanayake, Scott E. Fraser, Tatjana Sauka-Spengler. 2017. Biotagging of Specific Cell Populations in Zebrafish Reveals Gene Regulatory Logic Encoded in the Nuclear Transcriptome. Cell Reports Volume 19, Issue 2, p425–440
Posted by Kota Miura, on 15 May 2017
 NEUBIAS, the Network of European
NEUBIAS, the Network of European
BioImage Analysts (www.neubias.org), is delighted to announce two new Training Schools on BioImage Analysis:
The focus of training will be on construction and automation of image analysis workflows, using as examples more than one toolbox and different exercises. The schools will be held in Gothenburg 11-14th of September 2017, hosted by the Centre for Cellular Imaging – Sahlgrenska Academy, University of Gothenburg, Sweden.
NEUBIAS schools are an excellent opportunity to learn from many experts in bioimage analysis (we are expecting ~40 specialists at the event) and “….a great mix of intensive learning and community networking” (former trainee testimonial!).
More information about schools (programme & trainers) and venue, travel & lodge available at our website (linked above).
On behalf of all NEUBIAS members,
Julien Colombelli, Chair; Kota Miura, Vice-Chair
Julia Fernandez-Rodriguez, Local organizer
Carolina Wählby, Jan Eglinger, Joakim Lindblad & Nuno P Martins, TS4&5 programme organizers
Gaby G Martins & Fabrice Cordelières, WG2-Training leaders
Perrine Paul-Gilloteaux, WG4-Webtool leader
Sébastien Tosi, WG5-Benchmarking & Sample Datasets leader
NEUBIAS is an European network of currently ~180 members and 35 countries, which aims to promote the communication between Life Scientists, Instrumentalists, Developers and BioImage Analysts and to establish and promote the role of Bioimage Analysts in Life Science. Our mission includes:
Posted by the Node Interviews, on 11 May 2017
Our 20th instalment of this series comes from South Korea and features an investigation into the molecular basis of how temperature influences developmental transitions in Arabidopsis seedlings, recently published in Developmental Cell. We caught up with joint first authors Jun-Ho Ha and Hyo-Jun Lee, and their supervisor Chung-Mo Park, Professor in the Department of Chemistry, Seoul National University (SNU), to hear the story of the paper.

CMP I am currently professor in the Department of Chemistry at SNU. I earned my Bachelor of Science in the Department of Science Education from Seoul National University in 1983 and my PhD in molecular virology from State University of New York at Buffalo in 1993 under the supervision of professor Jeremy Bruenn. The topic of my thesis work was identification of killer toxin genes in a double-stranded virus endogenously residing in Ustilago maydis, a corn smut fungus and functional and structural characterization of the killer toxin proteins. After completing my PhD, I worked as a postdoctoral researcher in the same university and the Hauptman-Woodward Medical Research Institute, Buffalo, until I joined the Kumho Life & Environmental Science Laboratory, Korea, as PI in 1996. In the Kumho Laboratory, I worked on the photochemical and photobiological characterization of phytochrome photoreceptors in higher plants and the cyanobacteria Synechocystis PCC6803 and their associated light signal transduction in photomorphogenic responses.
In 2002, I accepted an associate professor position in the Department of Chemistry, SNU, where I have been since that time. While at SNU, my research team has been working on diverse aspects of plant growth and developmental processes, such as seed germination, phase transition and flowering induction, and leaf senescence. I have also been working on plant responses to environmental stresses with emphasis on temperature extremes and drought stresses. In recent years, my research is focused on plant adaptation to high but nonstressful temperatures (warm temperatures) with emphasis on leaf hyponasty, heat dissipation from leaves, and autotrophic development.
CMP The Korean government and several biotech companies have been investing a huge amount of research fund during the last 30 years. While industrial research and development has been a priority as a potential driving force of economic growth, the Korean government is also spending heavily on basic research. In plant science, there is a national research supporting program, termed New-Generation Biogreen 21, which is organized and supported by the Korean Rural Development Administration. The Program supports various research on both model plants and crops. It is considered that although not sufficient, enthusiastic plant scientists are able to get enough research funds to perform both basic and applied researches in recent years.
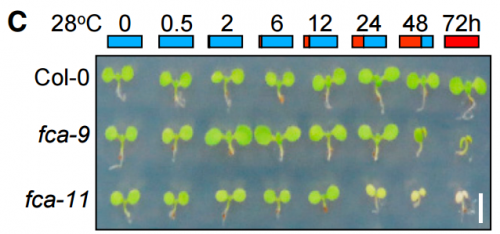
JHH I earned my Bachelor of Science in chemistry. I was also interested in molecular biology with an expectation that combining chemical and biological principles would be exciting in understanding life. While I was looking for an appropriate lab for my graduate study, I met Chung-Mo Park, who is my current thesis advisor. I was greatly impressed by his passion for science and research. It was also impressive that his group is working on plant molecular biology in the Department of Chemistry. I therefore decided to join his laboratory for my graduate study.
HJL Since I was a high school student, I planned to be a scientist with an aim of discovering unknown principles of nature and living organisms. After I entered the Department of Chemistry, Seoul National University, as an undergraduate student, I searched for potential labs in the Department appropriate for my research carrier. I realized that Chung-Mo’ lab is unique among the laboratories in that he is studying plant molecular biology and biochemistry. I thought that understanding molecular biological and biochemical mechanisms underlying plant performance would be helpful for me to find ways to sustain the Earth’s ecosystem. In particular, as a chemist, I thought that applying chemical tools to understanding biological systems would be interesting. I therefore decided to perform my graduate study in his lab.
CMP, JHH & HJL It is well known that extreme temperatures significantly affect plant performance, including autotrophic development. In addition, associated molecular events and signaling schemes are fairly well understood. In nature, the soil temperature is rapidly elevated under warm temperature conditions. Therefore, developing seedlings should cope with high temperatures while they pass through the heat-absorbing soil layer to obtain photosynthetic capacity required for autotrophic growth. However, it is almost unknown how the heat-labile shoot apical meristem tissues of developing seedlings handle the temperature constraints. It has recently been reported that warm temperatures, in a temperature range of 23 – 28oC in Arabidopsis, accelerate cell elongation during early seedling development. Thus, we were curious about whether and how warm temperatures influence chlorophyll biosynthesis during autotrophic development.
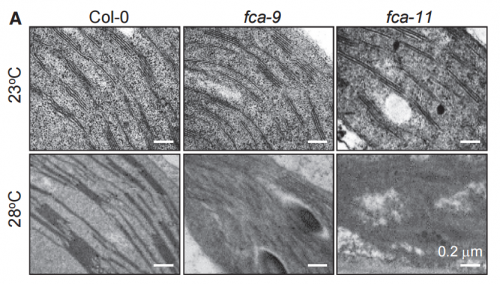
CMP, JHH & HJL We demonstrated that developing seedlings are capable of maintaining chlorophyll biosynthesis required for autotrophic development at warm temperature conditions. A group of photooxidoreductase (POR) enzymes is responsible for chlorophyll biosynthesis. Notably, they are susceptible to warm temperatures and thus rapidly inactivated in developing seedlings while they pass through the warm soil layer. We found that an RNA-binding protein FCA maintains the abundance of POR enzymes at warm temperatures in developing seedlings. Without FCA, plants fail to maintain the enzyme abundance, resulting in loss of chlorophyll and thus failure to achieve autotrophic growth. Our work provide a molecular basis for the acquisition of autotrophic growth under fluctuating temperature conditions in plants.
CMP, JHH & HJL Our recent findings strongly support that the typical RNA-binding protein FCA plays a critical role through epigenetic control of target genes during high temperature responses and thermomorphogenesis in Arabidopsis. Our data also indicate that FCA sustains the thermos-stable expression of POR enzymes during autotrophic development at warm temperatures. Altogether, these observations suggest that FCA function is thermos-regulated. However, it is current unclear how FCA is activated by ambient temperatures. We found that gene transcription and protein stability of FCA are not altered by temperature changes. Its subcellular localization is also unaltered under fluctuating temperature conditions.
Our preliminary data suggest that warm temperatures activates FCA through post-translational modifications, such as protein phosphorylation. We are currently under way to examine if FCA is differentially phosphorylated or chemically modified in response to temperature changes by employing global-scale proteomics.
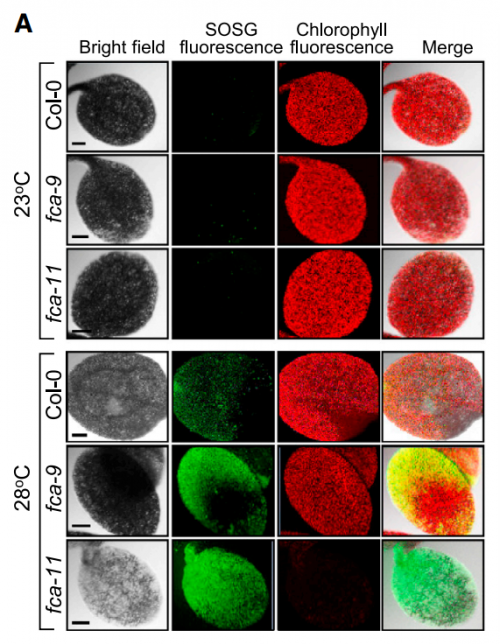
CMP, JHH & HJL Global warming depicts the gradual elevation of the average temperature of the Earth’s climate system. It is widely documented that under high ambient temperature conditions, plants exhibit distinct morphological and developmental traits, such as accelerated hypocotyl growth, leaf hyponasty, reduction of stomatal density, and early flowering, which profoundly influence crop productivity and commercial values. Our findings on plant thermal responses are closely associated with global warming. We propose that the FCA-mediated thermal adaptation of autotrophic development allows developing seedlings to cope with the heat-absorbing soil surface layer under natural conditions. In particular, we found that a single gene mutation causes a total loss of chlorophyll biosynthesis and autotrophic development at warm temperatures, providing a way of enhancing plant adaptation to thermal fluctuations in crop agriculture.
HJL & JHH In the initial stage of the research, we germinated and grew the FCA-defective mutants at normal temperatures for 3 days before transferred to warm temperatures to see if the fca mutations affect seedling growth. However, we did not observe any phenotypic differences in seedling growth and greening patterns in the mutants. A few months later, we anticipated that the fca mutations might affect the earlier stages of seedling growth. To examine the hypothesis, we germinated and grew the mutant seedlings at 28oC. We were surprised at the albino phenotype of the mutants. This observation triggered the re-examination of the thermal phenotypes of the fca mutants, resulting in the completion of this paper.
At first, we could not figure out why the fca mutants exhibited albinism only when germinated and grown at warm temperatures. As a potential cause of the albino phenotype, we considered several possibilities, such as defects in chloroplast development, chlorophyll biosynthesis, or both. It was found that the expression of POR genes was disrupted in the fca mutants when grown at warm temperatures. Accordingly, the level of chlorophylls was extremely low in the mutants, showing that the thermo-sensitive albino phenotype of the mutants is caused primarily by defects in chlorophyll biosynthesis, consistent with the FCA-mediated stabilization of POR production.
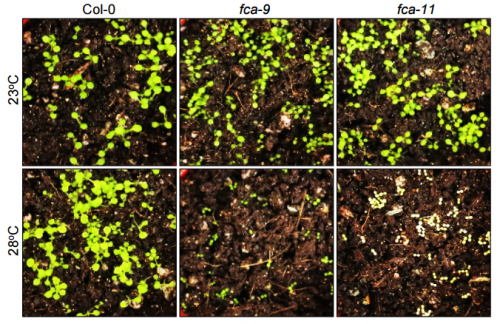
HJL & JHH The FCA-defective mutants are well-known late flowering mutants. A set of transgenic fca plants expressing POR genes were required for this study. It needs a lot of time to generate the transgenic plants because it takes 3-4 months to obtain seeds from the transgenic plants. While we were generating transgenic plants, we realized that a wrong expression construct was accidentally used, spending at least 5 additional months to obtain correct transgenic plants.
We also remember the frustrating moment when temperature controllers in the culture room were out of order during last summer, when we experienced a rarely high temperature and thus unstable supply of electricity in Korea. We had to grow a full set of plants again after a period time for fixing the temperature controllers.
HJL I am currently a postdoc in Chung-Mo Park’s lab. I will continue studying for a while on molecular and physiological mechanisms underlying plant thermomorphogenesis. I am interested in the as-yet unidentified regulator of POR abundance at warm temperatures. After finishing the experiments, I am planning to find an appropriate postdoc position to extend my research career in environmental control of plant proteomics.
JHH I hope to be able to finish my thesis study in a couple of years, after which I am planning to find postdoc positions in Korea or in USA to extend my research career in the field.
CMP We have a well-organized research system with a variety of molecular and biochemical tools, personnel, and facilities. We are specialized in gene regulatory mechanisms with emphasis on induction and activation mechanisms of transcription factors. Using these research tools and system, we will further extend our researches on plant thermomorphogenesis, which is emerging as a hot issue in the field because of the growing concern about global warming. In particular, we are focused on the functional linkage between photomorphogenic responses and growth hormones. We are also preparing a long-term project for engineering crop plants to enhance their adaptation capacity to changing temperature environment.
Jun-Ho Ha, Hyo-Jun Lee, Jae-Hoon Jung and Chung-Mo Park. 2017. Thermo-Induced Maintenance of Photo-oxidoreductases Underlies Plant Autotrophic Development. Developmental Cell 41(2): 170-179.
Browse the People behind the Papers archive here
Posted by The Francis Crick Institute, on 11 May 2017
Closing Date: 15 March 2021
Location: The Francis Crick Institute, Midland Road, London
Contract: Fixed-term (3 years), Full time
Salary: Competitive with benefits, subject to skills and experience
Vacancy ID: 5003
SHORT INTRODUCTION/SUMMARY
We seek a talented and motivated postdoc to join a Research Group led by Victor Tybulewicz at the Francis Crick Institute. The Group currently consists of 12 scientists, including 6 postdocs and 4 PhD students. One of the two main research interests of the Group is the study of the genetics underlying Down Syndrome. The Group has previously generated a series of mouse models of Down Syndrome that can be used to map the location of dosage-sensitive genes that cause Down Syndrome phenotypes (Lana-Elola et al, eLife 2016).
PROJECT SCOPE/ DESCRIPTION
The postdoc will study the genetics and developmental biology underlying congenital heart defects in Down Syndrome. The overall aim is to understand how increased dosage of genes on human chromosome 21 leads to heart defects. Specifically, the project aims to identify the dosage-sensitive genes that cause heart defects when present in three copies and to elucidate the mechanism by which the genes cause pathology. The work will involve use of genetic, developmental biology and biochemical techniques including microscopy, image analysis, and RNAseq, and will be supported by the excellent core facilities of the Institute. The work is funded by the Wellcome Trust.
The Francis Crick Institute is a biomedical discovery institute dedicated to understanding the fundamental biology underlying health and disease. Its work is helping to understand why disease develops and to translate discoveries into new ways to prevent, diagnose and treat illnesses such as cancer, heart disease, stroke, infections, and neurodegenerative diseases.
An independent organisation, its founding partners are the Medical Research Council (MRC), Cancer Research UK, Wellcome, UCL (University College London), Imperial College London and King’s College London.
The Crick was formed in 2015, and in 2016 it moved into a brand new state-of-the-art building in central London which brings together 1500 scientists and support staff working collaboratively across disciplines, making it the biggest biomedical research facility under a single roof in Europe.
The Francis Crick Institute will be world-class with a strong national role. Its distinctive vision for excellence includes commitments to collaboration; to developing emerging talent and exporting it the rest of the UK; to public engagement; and to helping turn discoveries into treatments as quickly as possible to improve lives and strengthen the economy.
If you are interested in applying for this role, please apply via our website https://goo.gl/IaFC2r
The closing date for applications is 10 June at 23:30 pm.
Please note: all offers of employment are subject to successful security screening and continuous eligibility to work in the United Kingdom.
Posted by Bridget Samuels, on 9 May 2017
We are pleased to announce the Center for Dental, Oral, Craniofacial Tissue and Organ Regeneration (C-DOCTOR – www.c-doctor.org) RFP that will award funding to promising dental, oral and craniofacial tissue engineering and regenerative medicine technologies and help them advance toward human clinical trials through customized product development advice and core resources. Please see the full RFP below the cut or here for details – deadline June 9, 2017. We ask that you kindly distribute this RFP widely to investigators who may be interested.
Posted by the Node, on 9 May 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. See June’s introductory post for background, and let us know if we missed anything
At the end of April, life science preprinting received a boost with the news that the Chan Zuckerberg Initiative will provide funding for the main preprint server, bioRxiv, which had the week before celebrated hosting its 10,000th article.
bioRxiv just passed 10,000 papers! :-)
Huge thanks to all involved https://t.co/xfNeiSBk3v pic.twitter.com/BQhIohafNX— Richard Sever (@cshperspectives) April 21, 2017
And a good month for bioRxiv was also a good month for developmental biology (and related) preprints. This month we found 115 preprints covering coral regeneration, spider development, a root-on-a-chip, a lot of discussion about publishing in our ‘Research Practise’ section, and a range of stem cell and cell biology, hosted on bioRxiv, F1000Research, PeerJ and arXiv.
Use these links to get to the section you want –
| Stem cells, regeneration & disease modelling
An Embryonic System To Assess Wnt Transcriptional Targets. Jahnavi Suresh, Nathan Harmston, Ka Keat Lim, Prameet Kaur, Helen Jingshu Jin, Jay B. Lusk, Enrico Petretto, Nicholas S. Tolwinski
C. elegans Flavin Monooxygenases Regulate C. elegans Axon Guidance and Growth Cone Protrusion with UNC-6/Netrin signaling and Rac GTPases. Mahekta R. Gujar, Aubrie M. Stricker, Erik A. Lundquist
TGF(beta) Mediated Structural Remodeling Facilitates Optic Fissure Fusion And The Necessity Of BMP Antagonism In This Process. Max D. Knickmeyer, Juan L. Mateo, Priska Eckert, Eleni Roussa, Belal Rahhal, Aimee Zuniga, Kerstin Krieglstein, Joachim Wittbrodt, Stephan Heermann
Alternative cleavage of a bone morphogenetic protein (BMP) produces ligands with distinct developmental functions and receptor preference. Edward N Anderson, Kristi A Wharton
MCAM controls cell autonomous polarity in myogenic and chondrogenic differentiation. Artal Moreno-Fortuny, Laricia Bragg, Giulio Cossu, Urmas Roostalu
The Calcineurin-FoxO-MuRF1 Signaling Pathway Regulates Myofibril Integrity in Cardiomyocytes. Jau-Nian Chen, Hirohito Shimizu, Adam D Langenbacher, Jie Huang, Kevin Wang, Georg Otto, Robert Geisler, Yibin Wang
Secretogranin-II Plays A Critical Role In Zebrafish Neurovascular Modeling. Binbin Tao, Hongling Hu, Kimberly Mitchell, Ji Chen, Haibo Jia, Zuoyan Zhu, Vance Trudeau, Wei Hu
Sympathetic Nerve Activity Promotes Cardiomyocyte Cell-Cycle Arrest And Binucleation. Li Chen, Alexander Y Payumo, Kentaro Hirose, Rachel B. Bigley, Jonathan Lovas, Rejji Kuruvilla, Guo N. Huang
Par3/Baz levels control epithelial folding at actomyosin-enriched compartmental boundaries. Jose M Urbano, Huw W Naylor, Elena Scarpa, Leila Muresan, Bénédicte Sanson
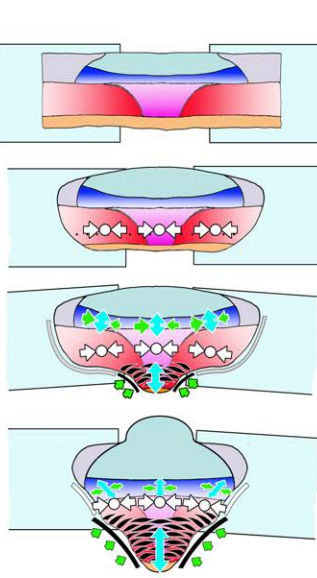
Large, long range tensile forces drive convergence during Xenopus blastopore closure and body axis elongation. David R Shook, Raymond Keller, Lance Davidson, Eric M. Kasprowicz
Cells From The Same Lineage Switch From Reduction To Enhancement Of Size Variability In Arabidopsis Sepals. Satoru Tsugawa, Nathan Hervieux, Daniel Kierzkowski, Anne-Lise Routier-Kierzkowska, Aleksandra Sapala, Olivier Hamant, Richard S. Smith, Adrienne H. K. Roeder, Arezki Boudaoud, Chun-Biu Li
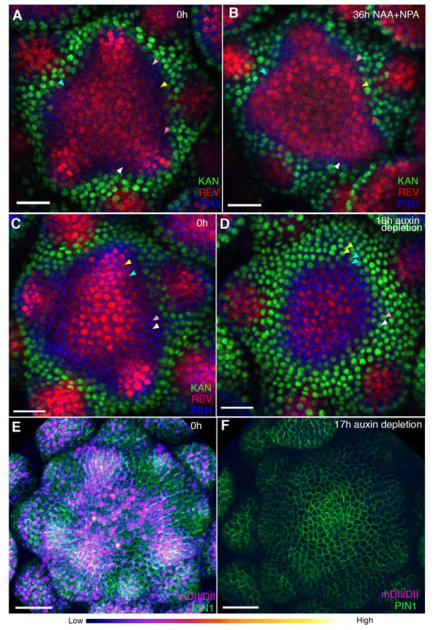
Cell type boundaries organize plant development. Monica Pia Caggiano, Xiulian Yu, Neha Bhatia, André Larsson, Hasthi Ram, Carolyn K Ohno, Pia Sappl, Elliot M Meyerowitz, Henrik Jönsson, Marcus G Heisler
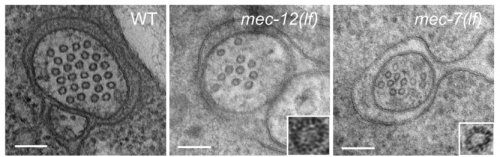
Distinct effects of tubulin isotype mutations on neurite growth in Caenorhabditis elegans. Chaogu Zheng, Margarete Diaz-Cuadros, Susan Laura Jao, Ken Nguyen, David H Hall, Martin Chalfie
Myomerger Induces Fusion Of Non-Fusogenic Cells And Is Required For Myoblast Fusion. Malgorzata Quinn, Qingnian Goh, Mitsutoshi Kurosaka, Dilani Gamage, Michael Petrany, Vikram Prasad, Douglas Millay
Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Shai Joseph, Mate Palfy, Lennart Hilbert, Mukesh Kumar, Jens Karschau, Vasily Zaburdaev, Andrej Shevchenko, Nadine Vastenhouw
Comprehensive Characterization Of The Complex Lola Locus Reveals A Novel Role In The Octopaminergic Pathway Via Tyramine Beta-Hydroxylase Activation. Nadja Dinges, Violeta Morin, Nastasja Kreim, Tony Southall, Jean-Yves Roignant
Genomic and chromatin features shaping meiotic double-strand break formation and repair in mice. Shintaro Yamada, Seoyoung Kim, Sam E Tischfield, Julian Lange, Maria Jasin, Scott Keeney
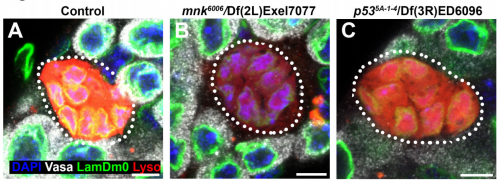
Germ cell connectivity enhances cell death in response to DNA damage in Drosophila testis. Yukiko M Yamashita, Kevin L Lu
The G-box transcriptional regulatory code in Arabidopsis. Daphne Ezer, Samuel JK Shepherd, Anna Brestovitsky, Patrick Dickinson, Sandra Cortijo, Varodom Charoensawan, Mathew S Box, Surojit Biswas, Philip Wigge
Cross-talk between active DNA demethylation, resetting of cellular metabolism and shoot apical growth in poplar bud break. Daniel Conde, Mariano Perales, Anne-Laure Le Gac, Christopher Dervinis, Matias Kirst, Stephane Maury, Pablo Gonzalez-Melendi, Isabel Allona

A Role For The F-Box Protein HAWAIIAN SKIRT In Plant miRNA Function. Patricia Lang, Michael Christie, Ezgi Dogan, Rebecca Schwab, Joerg Hagmann, Anna-Lena Van de Weyer, Detlef Weigel
Sequence Features Of MADS-Domain Proteins That Act As Hubs In The Protein-Protein Interaction Network Controlling Flower Development. Florian Ruempler, Guenter Theissen, Rainer Melzer
Nuclear Microenvironments Modulate Transcription From Low-Affinity Enhancers. Justin Crocker, Albert Tsai, Anand K Muthusamy, Luke D Lavis, Robert H Singer, David L Stern
Genetical genomics reveals Ras/MAPK modifier loci. Mark G. Sterken, Linda Van Bemmelen van der Plaat, Joost A.G. Riksen, Miriam Rodriguez, Tobias Schmid, Alex Hajnal, Jan E. Kammenga, Basten L. Snoek
5-Hydroxymethylcytosine Is Highly Dynamic Across Human Fetal Brain Development. Helen Spiers, Eilis Hannon, Leonard Schalkwyk, Nicholas Bray, Jonathan Mill
OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. Joel Hrit, Cheng Li, Elizabeth Allene Martin, Mary Goll, Barbara Panning
Transcription Activation Of Early Human Development Suggests DUX4 As An Embryonic Regulator. Virpi Töhönen, Shintaro Katayama, Liselotte Vesterlund, Mona Sheikhi, Liselotte Antonsson, Giuditta Filippini-Cattaneo, Marisa Jaconi, Anna Johnsson, Sten Linnarsson, Outi Hovatta, Juha Kere
Morphogen And Community Effects Determine Cell Fates In Response To BMP4 Signaling In Human Embryonic Stem Cells. Anastasiia Nemashkalo, Albert Ruzo, Idse Heemskerk, Aryeh Warmflash
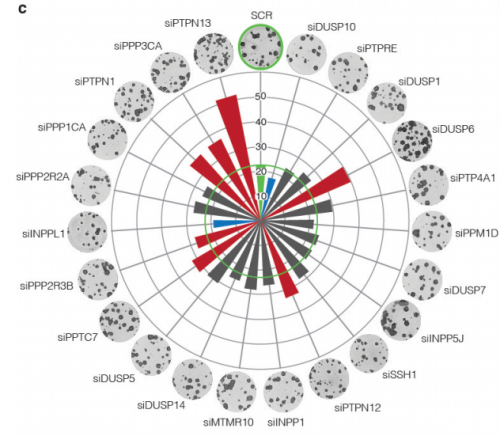
A protein phosphatase network controls temporal and spatial dynamics of differentiation commitment in human epidermis. Ajay Mishra, Angela Oliveira Pisco, Benedicte Oules, Tony Ly, Kifayathullah Liakath-Ali, Gernot Walko, Priyalakshmi Viswanathan, Jagdeesh Nijjher, Sara-Jane Dunn, Angus I Lamond, Fiona M Watt
The role of Cdx2 as a lineage specific transcriptional repressor for pluripotent network during trophectoderm and inner cell mass specification. Daosheng Huang, Xiaoping Han, Ping Yuan, Amy Ralston, Lingang Sun, Mikael Huss, Tapan Mistri, Luca Pinello, Huck Hui Ng, Guocheng Yuan, Junfeng Ji, Janet Rossant, Paul Robson, Guoji Guo
A lncRNA/Lin28/Let7 Axis Coupled To DNA Methylation Fine Tunes The Dynamics Of A Cell State Transition. Meng Amy Li, Paulo P. Amaral, Priscilla Cheung, Jan H. Bergmann, Masaki Kinoshita, Tuzer Kalkan, Meryem Ralser, Sam Robson, Ferdinand von Meyenn, Maike Paramor, Fengtang Yang, Caifu Chen, Jennifer Nichols, David L. Spector, Tony Kouzarides, Lin He, Austin Smith
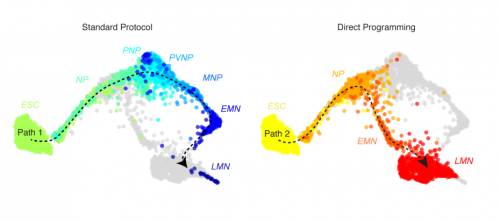
Mouse embryonic stem cells can differentiate via multiple paths to the same state. James Alexander Briggs, Victor C Li, Seungkyu Lee, Clifford J Woolf, Allon Klein, Marc W Kirschner
Pervasive Discordance Between mRNA And Protein Expression During Embryonic Stem Cell Differentiation. Patrick van den Berg, Bogdan Budnik, Nikolai Slavov, Stefan Semrau
Establishment In Culture Of Expanded Potential Stem Cells. Jian Yang, David Ryan, Wei Wang, Cheuk-Ho J Tsang, Guocheng Lan, Xuefei Gao, Liliana Antunes, Adam Clifford Wilkinson, Yong Yu, Aleksandra Kolodziejczyk, Lia Campos, Juexuan Wang, Fengtang Yang, Yosuke Tanaka, Melanie Eckersley-Maslin, Michael Woods, James Bussell, Ramiro Ramirez-Solis, Wolf Reik, Bertie Gottgens, Xiangang Zou, Liming Lu, Cui Wang, Hideki Masaki, Jacqui White, Hiro Nakauchi, Zheng Zhong, Sarah Teichmann, Beiyuan Fu, Zhexin Zhu, Pentao Liu
Fused dorsal-ventral cerebral organoids model human cortical interneuron migration. Joshua A Bagley, Daniel Reumann, Shan Bian, Juergen A. Knoblich
Transcription Factors Orchestrate Dynamic Interplay Between Genome Topology And Gene Regulation During Cell Reprogramming. Ralph Stadhouders, Enrique Vidal, François Serra, Bruno Di Stefano, François Le Dily, Javier Quilez, Antonio Gomez, Samuel Collombet, Clara Berenguer, Yasmina Cuartero, Jochen Hecht, Guillaume Filion, Miguel Beato, Marc A. Marti-Renom, Thomas Graf
Direct Conversion Of Human Fibroblasts Into Osteoblasts And Osteocytes With Small Molecules And A Single Factor, Runx2. Yanjiao Li, YaoLong Wang, Juehua Yu, Zhaoxia Ma, Qiong Bai, Xingfei Wu, Pengfei Bao, Lirong Li, Daiping Ma, Jingxue Liu, Change Liu, Fangyun Chen, Min Hu
RNA helicase, DDX27 regulates proliferation and myogenic commitment of muscle stem cells. Alexis Bennett, Marie Francoise O’Donohue, Stacey Gundry, Aye Chan, Jeffery Widrick, Isabelle Draper, Anirban Chakraborty, Yi Zhou, Leonard Zon, Pierre-Emmanuel Gleizes, Alan Beggs, Vandana Gupta
Constitutive Immune Activity Promotes Tumorigenesis in Drosophila Intestinal Progenitor Cells. Kristina Petkau, Silvia Guntermann, Edan Foley
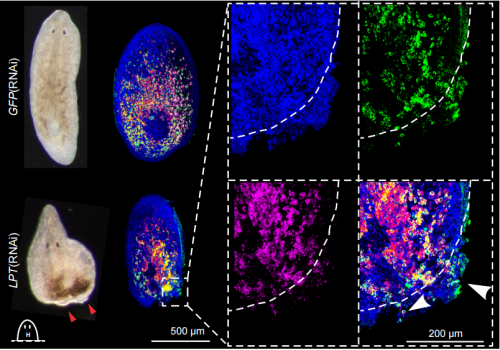
Conservation of EMT transcription factor function in controlling pluripotent adult stem cell migration in vivo in planarians. Prasad Abnave, Ellen Aboukhatwa, Nobuyoshi Kosaka, James Thompson, Mark Hill, Aziz Aboobaker
MLL3/4 Prevents Stem Cell Hyperplasia And Controls Differentiation Programs In A Planarian Cancer Stem Cell Model. Yuliana Mihaylova, Damian Kao, Samantha Hughes, Alvina Lai, Farah Jaber-Hijazi, Nobuyoshi Kosaka, Prasad Abnave, Aziz Aboobaker
PHRED-1 Is A Divergent Neurexin-1 Homolog That Organizes Muscle Fibers And Patterns Organs During Regeneration. Carolyn E. Adler, Alejandro Sanchez Alvarado
Post-transcriptional regulation of adult CNS axonal regeneration by Cpeb1. Wilson Pak-Kin Lou, Alvaros Mateos, Marta Koch, Stefan Klussmann, Chao Yang, Na Lu, Stefanie Limpert, Manuel Göpferich, Marlen Zschaetzsch, Carlos Maillo, Elena Senis, Dirk Grimm, Raúl Méndez, Kai Liu, Bassem A Hassan, Ana Martin-Villalba
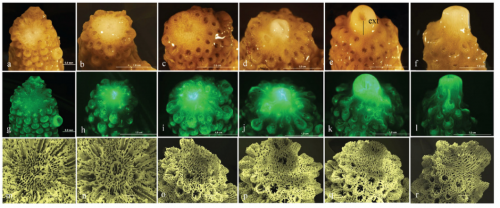
Exposure to elevated sea-surface temperatures below the bleaching threshold impairs coral recovery and regeneration following injury. Joshua Louis Boness, William Leggat, Tracy Danielle Ainsworth
Necroptosis promotes the Aging of the Male Reproductive System in Mice. Xiaodong Wang, Dianrong Li, Lingjun Meng, Tao Xu, Yaning Su, Xiao Liu, Zhiyuan Zhang
Tissue-specific downregulation of EDTP removes polyglutamine protein aggregates and extends lifespan in Drosophila. Chengfeng Xiao, Shuang Qiu, R Meldrum Robertson, Laurent Seroude
Reversal of cardiac and skeletal manifestations of Duchenne muscular dystrophy by cardiosphere-derived cells and their exosomes in mdx dystrophic mice and in human Duchenne cardiomyocytes. Mark A Aminzadeh, Russell G Rogers, Kenneth Gouin, Mario Fournier, Rachel E Tobin, Xuan Guan, Martin K Childers, Allen M Andres, David J Taylor, Ahmed Ibrahim, Xiang-ming Ding, Angelo Torrente, Joshua I Goldhaber, Ronald A Victor, Roberta A Gottlieb, Michael Lewis, Eduardo Marban
PERTURBATION OF PTEN-PI3K/AKT SIGNALLING IMPAIRED AUTOPHAGY MODULATION IN DYSTROPHIN-DEFICIENT MYOBLASTS. Muhammad Dain Yazid, Janet Smith
Modeling Zika Virus Congenital Eye Disease: Differential Susceptibility of Fetal Retinal Progenitor Cells and iPSC-Derived Retinal Stem Cells to Zika Virus Infection. Deisy Contreras, Melissa Jones, Laura E Martinez, Vineela Gangalapudi, Jie Tang, Ying Wu, Jiagang J. Zhao, Zhaohui Chen, Shaomei Wang, Vaithilingaraja Arumugaswami
Astral microtubule dynamics regulate anaphase oscillation onset and set a robust final position for the Caenorhabditis elegans zygote spindle. Helene Bouvrais, Laurent Chesneau, Sylvain Pastezeur, Marie Delattre, Jacques Pecreaux
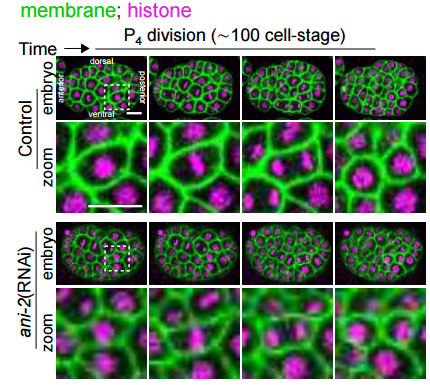
Anillin proteins stabilize the cytoplasmic bridge between the two primordial germ cells during C. elegans embryogenesis. Eugenie Goupil, Rana Amini, Jean-Claude Labbe
Local Nucleation Of Microtubule Bundles Through Tubulin Concentration Into A Condensed Tau Phase. Amayra Hernández-Vega, Marcus Braun, Lara Scharrel, Marcus Jahnel, Susanne Wegmann, Bradley T. Hyman, Simon Alberti, Stefan Diez, Anthony A. Hyman
An Arf6- And Caveolae-Dependent Pathway Links Hemidesmosome Remodeling And Mechanoresponse. Naël Osmani, Julien Pontabry, Jordi Comelles, Nina Fekonja, Jacky G Goetz, Daniel Riveline, Elisabeth Georges-Labouesse, Michel Labouesse
Cell size sensing in animal cells coordinates growth rates and cell cycle progression to maintain cell size uniformity. Miriam Bracha Ginzberg, Nancy Chang, Ran Kafri, Marc W Kirschner
Scc2-Mediated Loading Of Cohesin Onto Chromosomes In G1 Yeast Cells Is Insufficient To Build Cohesion During S Phase. Kim Nasmyth
Gradients Of Rac1 Nanoclusters Support Spatial Patterns Of Rac1 Signaling. Amanda Remorino, Simon De Beco, Fanny Cayrac, Fahima Di Federico, Gaetan Cornilleau, Alexis Gautreau, Maria Carla Parrini, Jean-Baptiste Masson, Maxime Dahan, Mathieu Coppey
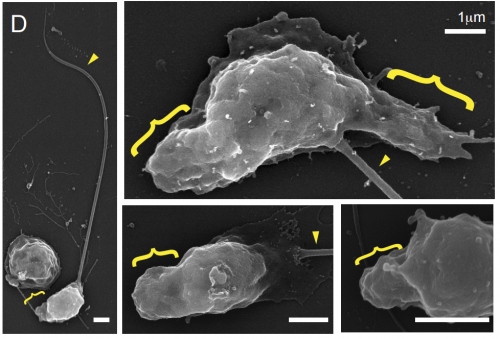
WASP and SCAR are evolutionarily conserved in actin-filled pseudopod-based motility. Lillian K. Fritz-Laylin, Samuel J. Lord, R. Dyche Mullins
Optogenetic Control of RhoA Reveals Zyxin-mediated Elasticity of Stress Fibers. Patrick W Oakes, Elizabeth Wagner, Christoph A Brand, Dimitri Probst, Marco Linke, Ulrich S Schwarz, Michael Glotzer, Margaret L Gardel
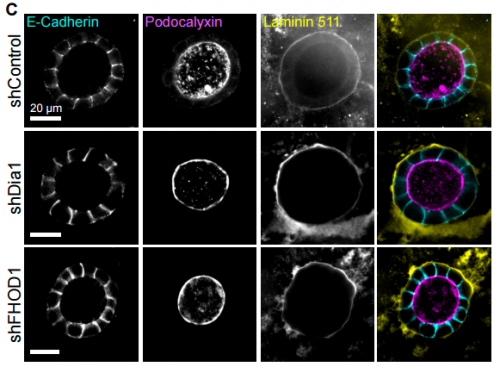
Formin-Dependent Adhesions Are Required For Invasion By Epithelial Tissues. Tim B Fessenden, Yvonne Beckham, Mathew Perez-Neut, Aparajita H Chourasia, Kay F Macleod, Patrick W Oakes, Margaret L Gardel
Fast Activation Cycles Of Rac1 At The Lamellipodium Tip Trigger Membrane Protrusion. Amine Mehidi, Olivier Rossier, Anael Chazeau, Fabien Biname, Amanda Remorino, Mathieu Coppey, Zeynep Karatas, Jean-Baptiste Sibarita, Violaine Moreau, Gregory Giannone
Combinatorial Regulation Of The Balance Between Dynein Microtubule End Accumulation And Initiation Of Directed Motility. Rupam Jha, Johanna Roostalu, Martina Trokter, Thomas Surrey
The Ndc80 complex targets Bod1 to human mitotic kinetochores. Katharina Schleicher, Sara ten Have, Iain M Porter, Jason R Swedlow
Dynactin Binding To Tyrosinated Microtubules Promotes Centrosome Centration In C. Elegans By Enhancing Dynein-Mediated Organelle Transport. Daniel J. Barbosa, Joana Duro, Dhanya K. Cheerambathur, Bram Prevo, Ana X. Carvalho, Reto Gassmann
Long-Term Memory In The Migration Movements Of Enucleated Amoeba proteus. Carlos Bringas, Iker Malaina, Alberto Perez-Samartin, Maria Dolores Boyano, Maria Fedetz, Gorka Perez-Yarza, Jesus Cortes, Ildefonso Martinez de la Fuente
Do gametes woo? Evidence for non-random unions at fertilization. Joseph H Nadeau
Pairwise hybrid incompatibilities dominate allopatric speciation for a simple biophysical model of development. Bhavin S Khatri, Richard Goldstein
A model for autonomous and non-autonomous effects of the Hippo pathway in Drosophila. Jia Gou, Lin Lin, Hans G Othmer
Single-Cell Genome Dynamics in Early Embryo Development: A Statistical Thermodynamics Approach. Alessandro Giuliani, Masa Tsuchiya, Kenichi Yoshikawa
On The Principles Of Cell Decision-Making: Intracellular Coupling Improves Cell Responses Fidelity Of Noisy Signals. Andreas Reppas, Eduard Jorswieck, Haralampos Hatzikirou
Correlating Cell Shape and Cellular Stress in Motile Confluent Tissues. Xingbo Yang, Dapeng Bi, Michael Czajkowski, Matthias Merkel, M. Lisa Manning, M. Cristina Marchetti
apterous A Specifies Dorsal Wing Patterns And Sexual Traits In Butterflies. Anupama Prakash, Antonia Monteiro
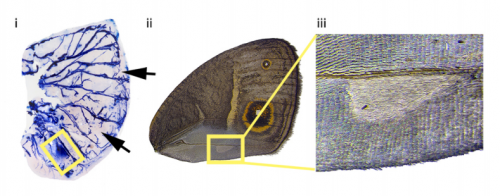
Sex Differences In 20-Hydroxyecdysone Hormone Levels Control Sexual Dimorphism In Bicyclus anynana Butterfly Wing Patterns. Shivam Bhardwaj, Kathleen L Prudic, Ashley Bear, Mainak Das Gupta, Bethany R Wasik, Xiaoling Tong, Wei Fun Cheong, Markus R Wenk, Antonia Monteiro
A practical guide to CRISPR/Cas9 genome editing in Lepidoptera. Linlin Zhang, Robert Reed
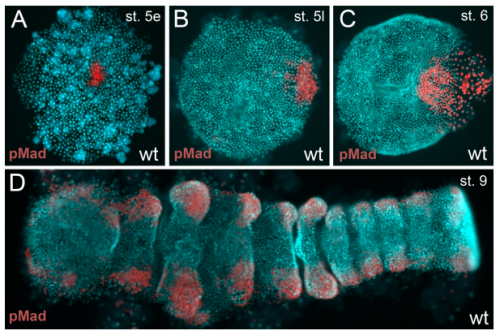
A novel role for Ets4 in axis specification and cell migration in the spider Parasteatoda tepidariorum. Matthias Pechmann, Matthew Alan Benton, Nathan James Kenny, Nico Posnien, Siegfried Roth
Evolution And Multiple Roles Of The Pancrustacea Specific Transcription Factor zelda In Insects. Lupis Ribeiro, Vitoria Tobias-Santos, Danielle Santos, Felipe Antunes, Georgia Feltran, Jackson de Souza Menezes, L Aravind, Thiago M Venancio, Rodrigo Nunes da Fonseca
Deep experimental profiling of microRNA diversity, deployment, and evolution across the Drosophila genus. Alex S Flynt, Alexandra Panzarino, Md Mosharrof Hossain Mondal, Adam Siepel, Jaaved Mohammed, Eric Lai
Single Molecule Fluorescence In Situ Hybridisation For Quantitating Post-Transcriptional Regulation In Drosophila Brains. Lu Yang, Josh Titlow, Darragh Ennis, Carlas Smith, Jessica Mitchell, Florence L. Young, Scott Waddell, David Ish-Horowicz, Ilan Davis
A Versatile Compressed Sensing Scheme For Faster And Less Phototoxic 3D Fluorescence Microscopy. Maxime Woringer, Xavier Darzacq, Christophe Zimmer, Mustafa Mir
Three-Dimensional Two-Photon Optogenetics And Imaging Of Neural Circuits In Vivo. Weijian Yang, Luis Carrillo-Reid, Yuki Bando, Darcy S. Peterka, Rafael Yuste
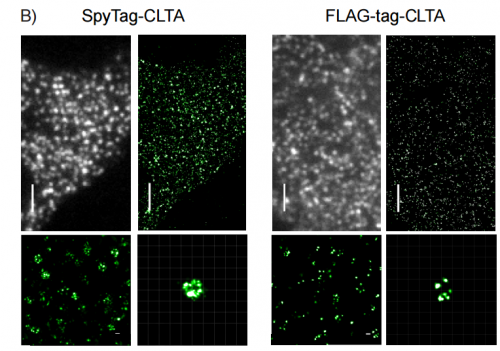
Covalent Protein Labeling By SpyTag-SpyCatcher In Fixed Cells For Super-Resolution Microscopy. Veronica Pessino, Y. Rose Citron, Siyu Feng, Bo Huang
Photoacoustic molecular rulers based on DNA nanostructures. James Joseph, Philipp Koehler, Tim J. Zuehlsdorff, Daniel J Cole, Kevin N. Baumann, Judith Weber, Sarah E. Bohndiek, Silvia Hernandez-Ainsa
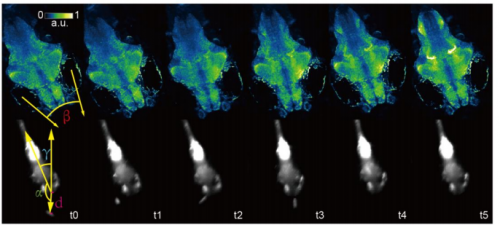
Rapid Whole Brain Imaging Of Neural Activities In Freely Behaving Larval Zebrafish. Lin Cong, Zeguan Wang, Yuming Chai, Wei Hang, Chunfeng Shang, Wenbin Yang, Lu Bai, Jiulin Du, Kai Wang, Quan Wen
A general method to fine-tune fluorophores for live-cell and in vivo imaging. Jonathan B. Grimm, Anand K. Muthusamy, Yajie Liang, Timothy A. Brown, William C. Lemon, Ronak Patel, Rongwen Lu, John J. Macklin, Phillip J. Keller, Na Ji,Luke D. Lavis
Real-Time Observation Of Light-Controlled Transcription In Living Cells. Anne Rademacher, Fabian Erdel, Jorge Trojanowski, Karsten Rippe
FiloQuant reveals increased filopodia density during DCIS progression. Guillaume Jacquemet, Ilkka Paatero, Alexandre Carisey, Artur Padzik, Jordan Orange, Hellyeh Hamidi, Johanna Ivaska
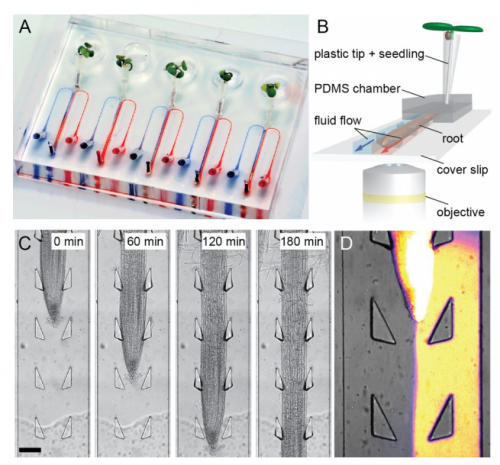
An organ-on-a-chip approach for investigating root-environment interactions in heterogeneous conditions. Claire E. Stanley, Jagriti Shrivastava, Rik Brugman, Dirk van Swaay, Guido Grossmann
Disabling Cas9 by an anti-CRISPR DNA mimic. Jiyung Shing, Fuguo Jiang, Jun-Jie Liu, Nicholas L Bray, Benjamin J Rauch, Seung Hyun Baik, Eva Nogales, Joseph Bondy-Denomy, Jacob E Corn, Jennifer A Doudna
Modulation of Genome Editing Outcomes by Cell Cycle Control of Cas9 Expression. Yuping Huang, Caitlin McCann, Andrey Samsonov, Dmitry Malkov, Greg D Davis, Qingzhou Ji
The WPRE Improves Genetic Engineering With Site-Specific Nucleases. Jessica M. Ong, Christopher R Brown, Matthew C. Mendel, Gregory J Cost
A Versatile Genetic Tool For Post-Translational Control Of Gene Expression With A Small Molecule In Drosophila melanogaster. Sachin Sethi, Jing W. Wang
Rapid DNA Re-Identification for Cell Line Authentication and Forensics. Sophie Zaaijer, Yaniv Erlich, Daniel Speyer, Robert Piccone, Assaf Gordon
Reading canonical and modified nucleotides in 16S ribosomal RNA using nanopore direct RNA sequencing. Andrew M Smith, Miten Jain, Logan Mulroney, Daniel R Garalde, Mark Akeson
Single-cell analysis of clonal dynamics in direct lineage reprogramming: a combinatorial indexing method for lineage tracing. Brent A Biddy, Sarah E Waye, Tao Sun, Samantha A Morris
Nanopore sequencing and assembly of a human genome with ultra-long reads. Miten Jain, Sergey Koren, Josh Quick, Arthur C Rand, Thomas A Sasani, John R Tyson, Andrew D Beggs, Alexander T Dilthey, Ian T Fiddes, Sunir Malla, Hannah Marriott, Karen H Miga, Tom Nieto, Justin O’Grady, Hugh E Olsen, Brent S Pedersen, Arang Rhie, Hollian Richardson, Aaron Quinlan, Terrance P Snutch, Louise Tee, Benedict Paten, Adam M. Phillippy, Jared T Simpson, Nicholas James Loman, Matthew Loose
Beyond The Linear Genome: Comprehensive Determination Of The Endogenous Circular Elements In C. elegans And Human Genomes Via An Unbiased Genomic-Biophysical Method. Massa Shoura, Idan Gabdank, Loren Hansen, Jason Merker, Jason Gotlib, Stephen Levene, Andrew Fire
Expression of short hairpin RNAs using the compact architecture of retroviral microRNA genes. James M Burke, Rodney P. Kincaid, Francesca Aloisio, Nicole Welch, Christopher S. Sullivan
Improved maize reference genome with single molecule technologies. Yinping Jiao, Paul Peluso, Jinghua Shi, Tiffany Liang, Michelle C Stitzer, Bo Wang, Michael Campbell, Joshua C Stein, Xuehong Wei, Chen-Shan Chin, Katherine Guill, Michael Regulski, Sunita Kumari, Andrew Olson, Jonathan Gent, Kevin L Schneider, Thomas K Wolfgruber, Michael R May, Nathan M Springer, Eric Antoniou, Richard McCombie, Gernot G Presting, Michael McMullen, Jeffrey Ross-Ibarra, R. Kelly Dawe, Alex Hastie, David R Rank, Doreen Ware
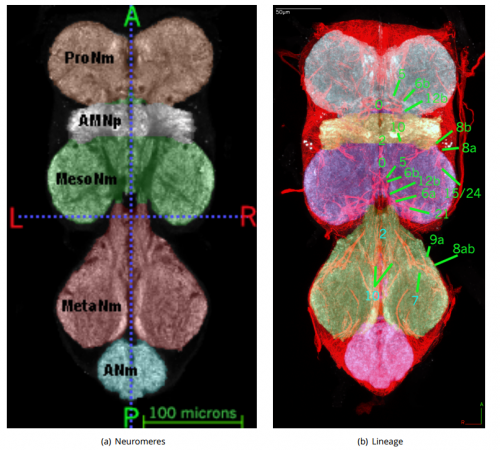
A Systematic Nomenclature for the Drosophila Ventral Nervous System. Robert Christopher Court, James Douglas Armstrong, Jana Borner, Gwyneth Card, Marta Costa, Michael Dickinson, Carsten Duch, Wyatt Korff, Richard Mann, David Merritt, Rod Murphey, Shigehiro Namiki, Andrew Seeds, David Shepherd, Troy Shirangi, Julie Simpson, James Truman, John Tuthill, Darren Williams
MAPseq: Improved Speed, Accuracy And Consistency In Ribosomal RNA Sequence Analysis. Joao F Matias Rodrigues, Thomas SB Schmidt, Janko Tackmann, Christian von Mering
High Accuracy Base Calls in Nanopore Sequencing. Philippe Christophe Faucon, Robert Trevino, Parithi Balachandran, Kylie Standage-Beier, Xiao Wang
BasePlayer: Versatile Analysis Software For Large-Scale Genomic Variant Discovery. Riku Katainen, Iikki Donner, Tatiana Cajuso, Eevi Kaasinen, Kimmo Palin, Veli Mäkinen, Lauri A Aaltonen, Esa Pitkänen
CiliaCarta: An Integrated And Validated Compendium Of Ciliary Genes. Teunis J. P. van Dam, Julie Kennedy, Robin van der Lee, Erik de Vrieze, Kirsten A. Wunderlich, Suzanne Rix, Gerard W. Dougherty, Nils J. Lambacher, Chunmei Li, Victor L. Jensen, Michael R. Leroux, Rim Hjeij, Nicola Horn, Yves Texier, Yasmin Wissinger, Jeroen van Reeuwijk, Gabrielle Wheway, Barbara Knapp, Jan F. Scheel, Brunella Franco, Dorus A. Mans, Erwin van Wijk, François Képès, Gisela G. Slaats, Grischa Toedt, Hannie Kremer, Heymut Omran, Katarzyna Szymanska, Konstantinos Koutroumpas, Marius Ueffing, Thanh-Minh T. Nguyen, Stef J. F. Letteboer, Machteld M. Oud, Sylvia E. C. van Beersum, Miriam Schmidts, Philip L. Beales, Qianhao Lu, Rachel H. Giles, Radek Szklarczyk, Robert B. Russell, Toby J. Gibson, Colin A. Johnson, Oliver E. Blacque, Uwe Wolfrum, Karsten Boldt, Ronald Roepman, Victor Hernandez-Hernandez, Martijn A. Huynen
Looking Into Pandora’s Box: The Content Of Sci-Hub And Its Usage. Bastian Greshake
Anticipated effects of an open access policy at a private foundation. Eesha Khare, Carly Strasser
Gender disparity in computational biology research publications. Kevin S. Bonham, Melanie I. Stefan
Why Do Scientists Fabricate And Falsify Data? A Matched-Control Analysis Of Papers Containing Problematic Image Duplications. Daniele Fanelli, Rodrigo Costas, Ferric C Fang, Arturo Casadevall, Elisabeth M Bik
Addressing the digital divide in contemporary biology: Lessons from teaching UNIX. Serghei Mangul, Lana Martin, Alexander Hoffmann, Matteo Pellegrini, Eleazar Eskin
The appropriation of GitHub for curation. Yu Wu, Na Wang, Jessica Kropczynski, John M Carroll
The earth is flat (p>0.05): Significance thresholds and the crisis of unreplicable research. Valentin Amrhein, Fränzi Korner-Nievergelt, Tobias Roth
What is open peer review? A systematic review. Tony Ross-Hellauer
Standardising and harmonising research data policy in scholarly publishing. Iain Hrynaszkiewicz,Aliaksandr Birukou, Mathias Astell, Sowmya Swaminathan, Amye Kenall, Varsha Khodiyar
TOWARDS COORDINATED INTERNATIONAL SUPPORT OF CORE DATA RESOURCES FOR THE LIFE SCIENCES. Warwick Anderson, Rolf Apweiler, Alex Bateman, Guntram A Bauer, Helen Berman, Judith A Blake, Niklas Blomberg, Stephen K Burley, Guy Cochrane, Valentina Di Francesco, Tim Donohue, Christine Durinx, Alfred Game, Eric Green, Takashi Gojobori, Peter Goodhand, Ada Hamosh, Henning Hermjakob, Minoru Kanehisa, Robert Kiley, Johanna McEntyre, Rowan McKibbin, Satoru Miyano, Barbara Pauly, Norbert Perrimon, Mark A Ragan, Geoffrey Richards, Yik-Ying Teo, Monte Westerfield, Eric Westhof, Paul F Lasko
Caterpillars lack a resident gut microbiome. Tobin J Hammer, Daniel H Janzen, Winnifred Hallwachs, Samuel L Jaffe, Noah Fierer
Estimating The Extinction Date Of The Thylacine Accounting For Unconfirmed Sightings. Colin J Carlson, Alexander L Bond, Kevin R Burgio
Posted by kwagner2, on 8 May 2017
Closing Date: 15 March 2021
A postdoctoral position is immediately available to pursue cutting-edge musculoskeletal stem cell research in the laboratory of Kathryn Wagner, MD, PhD at the Center for Genetic Muscle Disorders, Kennedy Krieger Institute and Johns Hopkins School of Medicine,
The Wagner laboratory is a moderate-size laboratory that focuses on translational science to develop novel therapies for muscle disorders. Approaches include small molecules, biologics, stem cells and gene therapy. The laboratory is part of a Senator Paul Wellstone Muscular Dystrophy Cooperative Research Center and has extensive collaboration both within Johns Hopkins and across institutions. https://www.thewagnerlab.org.
The ideal applicant will have a PhD in molecular or cell biology. Experience in skeletal muscle is a positive but not a requirement. This is a fully funded position to study transplantation of iPSC-derived myogenic progenitors in models of muscular dystrophy and volumetric muscle loss.
Interested applicants should send a cover letter and CV to Dr. Wagner at wagnerk@kennedykrieger.org