November in preprints
Posted by the Node, on 2 December 2016
Our latest monthly trawl for developmental biology (and other cool) preprints. See June’s introductory post for background, and let us know if we missed anything
This month, we found preprints covering various aspects of plant growth and patterning, a lot of cell biology – including insights into microtubules organisation, RNA localisation and yeast size control – as well as a bunch of tools. One of the most talked about preprints of the month comes from our ‘Away from the bench‘ section: a guide for how to structure scientific papers. Happy reading!
Developmental biology
SmallOrgan 1 plays an essential role in cell proliferation, cell expansion and cadmium uptake in rice. Peng Qin, Jiangbo Hu, Weilan Chen, Guohua Zhang, Jian Li, Shijun Fan, Bin Tu, Xuewei Chen, Yuping W Wang, Shigui Li, Bingtian Ma.
Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Hui Dong, Jack Dumenil, Fu-Hao Lu, Li Na, Hannes Vanhaeren, Cristin Naumann, Maria Klecker, Rachel Prior, Caroline Smith, Neil McKenzie, Gerhard Saalbach, Liangliang Chen, Tian Xia, Nathalie Gonzalez, Mathilde Seguela, Dirk Inze, Nico Dissmeyer, Yunhai Li, Michael W Bevan
Ancient coding sequences underpin the spatial patterning of gene expression in C4 leaves. Ivan Reyna-Llorens, Steven J Burgess, Ben P Williams, Susan Stanley, Chris Boursnell, Julian M Hibberd
The Tension-sensitive Ion Transport Activity of MSL8 is Critical for its Function in Pollen Hydration and Germination. Eric S. Hamilton, Elizabeth S. Haswell
tRNA-derived small RNAs target transposable element transcripts. Sarah Choudury, Keith Slotkin, German Martinez
Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Aaron C Daugherty, Robin Yeo, Jason D Buenrostro, William J Greenleaf, Anshul Kundaje, Anne Brunet
An RNA binding polymer specifies nematode sperm fate. Scott Takeo Aoki, Douglas F Porter, Aman Prasad, Marvin Wickens, Craig A Bingman, Judith Kimble
DIV-1/PolA2 Promotes GLP-1/Notch-Mediated Cellular Events in Caenorhabditis elegans. Dong Suk Yoon, Dong Seok Cha, Myon-Hee Lee
PRDX6 inhibits neurogenesis of neural precursor cells through downregulation of wdfy1 mediated TLR4 signal. Mi Hee Park, Dong Ju Son, Kyoung Tak Nam, So Young Kim, Sang Yeon Oh, Min Ji Song, Hyung Ok Chun, Tae Hyung Lee, Jin Tae Hong
Axonal transcriptome of human stem cell derived neurons. Rebecca L Bigler, Joyce W Kamande, Raluca Dumitru, Mark Niedringhaus, Anne Marion Taylor.
Estimating Drivers of Cell State Transitions Using Gene Regulatory Network Models. Daniel Schlauch, Kimberly Glass, Craig P Hersh, Edwin K Silverman, John Quackenbush
G protein-coupled estrogen receptor regulates heart rate in zebrafish embryos. Shannon N Romano, Hailey E Edwards, Jaclyn Paige Souder, Xiangqin Cui, Daniel A Gorelick
In vivo imaging of coral tissue and skeleton with optical coherence tomography. Daniel Wangpraseurt, Camilla Wentzel, Steven L Jacques, Michael Wagner, Michael Kuhl
Dynamic changes in Sox2 spatio-temporal expression direct the second cell fate decision through Fgf4/Fgfr2 signaling in preimplantation mouse embryos. Tapan Kumar Mistri, Wibowo Arindrarto, Wei Ping Ng, Choayang Wang, Hiong Lim Leng, Lili Sun, Ian Chambers, Thorsten Wohland, Paul Robson
High temporal resolution of gene expression dynamics in developing mouse embryonic stem cells. Brian S Gloss, Bethany Signal, Seth W Cheetham, Franziska Gruhl, Dominik Kaczorowski, Andrew C Perkins, Marcel E Dinger
Functional transcriptomics in diverse intestinal epithelial cell types reveals robust gut microbial sensitivity of microRNAs in intestinal stem cells. Bailey CE Peck, Amanda T Mah, Wendy A Pitman, Shengli Ding, P. Kay Lund, Praveen Sethupathy
Constitutive Immune Activity Promotes Tumorigenesis in Drosophila Intestinal Progenitor Cells. Kristina Petkau, Silvia Guntermann, Edan Foley
Rfx2 stabilizes Foxj1 binding at chromatin loops to enable multiciliated cell gene expression. Ian K Quigley, Chris Kintner
Ageing, TOR and amino acid restriction: a cross-tissue transcriptional network connects GATA factors to Drosophila longevity. Adam J Dobson, Xiaoli He, Eric Blanc, Ekin Bolukbasi, Yodit Feseha, Mingyao Yang, Matthew Piper
Pulse of α-2-macroglobulin and lipocalin-1 in the pregnant uterus of European polecats (Mustela putorius) at the time of implantation. Heli Lindeberg, Richard Burchmore, Malcolm W. Kennedy
Cell biology
A Critical-like Collective State Leads to Long-range Cell Communication in Dictyostelium discoideum Aggregation. Giovanna De Palo, Darvin Yi, Robert Endres
Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Eugene A Katrukha, Marina Mikhaylova, Hugo X van Brakel, Paul M van Bergen en Henegouwen, Anna Akhmanova, Casper C Hoogenraad, Lukas C Kapitein
Size-Dependent Accumulation of the Mitotic Activator Cdc25 as a Mechanism of Size Control in Fission Yeast. Daniel Keifenheim, Xi-Ming Sun, Edridge D’Souza, Makoto Ohira, Mira Magner, Michael B. Mayhew, Samuel Marguerat, Nicholas Rhind
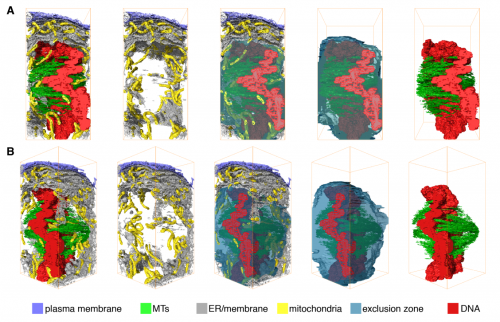
Microtubule organization within mitotic spindles revealed by serial block face scanning EM and image analysis. Faye M Nixon, Thomas R Honnor, Georgina P Starling, Alison J Beckett, Adam M Johansen, Julia A Brettschneider, Ian A Prior, Stephen J Royle
Microtubules with a twist: a lumenal interrupted helix in human sperm tail microtubules. John M Heumann, Cindi L Schwartz, Azusa Suzuki-Shinjo, Garry Morgan, Per Olov Widlund, Johanna Louise Hoog
Visualizing adenosine to inosine RNA editing in single mammalian cells. Ian A Mellis, Rohit K Gupte, Arjun Raj, Sara H Rouhanifard
An RNA-binding tropomyosin recruits kinesin-1 dynamically to oskar mRNPs. Imre Gaspar, Vasily Sysoev, Artem Komissarov, Anne Ephrussi
Drosophila BEACH domain autophagic adaptor blue cheese shuttles between vesicle populations and is required for an early step in autophagy. Joan Sim, Kathleen Amy Osborne, Irene Argudo Garcia, Artur Matysik, Rachel Kraut
Evolution, etc
Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in Drosophila. Anna B Buchman, Tobin Ivy, John M Marshall, Omar Akbari, Bruce A. Hay
Recurrent gene duplication leads to diverse repertoires of centromeric histones in Drosophila species. Lisa E. Kursel, Harmit Singh Malik
An annotated draft genome for Radix auricularia (Gastropoda, Mollusca). Tilman Schell, Barbara Feldmeyer, Hanno Schmidt, Bastian Greshake, Oliver Tills, Manuela Truebano, Simon D. Rundle, Juraj Paule, Ingo Ebersberger, Markus Pfenninger
Tools & Resources
PhysiCell: an Open Source Physics-Based Cell Simulator for 3-D Multicellular Systems. Ahmadreza Ghaffarizadeh, Samuel H. Friedman, Shannon M. Mumenthaler,Paul Macklin
pathVar: a new method for pathway-based interpretation of gene expression variability. Laurence de Torrente, Samuel Zimmerman, Deanne Taylor, Yu Hasegawa, Christine A Wells, Jessica C Mar
MultiCellDS: a community-developed standard for curating microenvironment-dependent multicellular data. Samuel H. Friedman, Alexander R.A. Anderson, David M. Bortz, Alexander G. Fletcher, Hermann B. Frieboes, Ahmadreza Ghaffarizadeh, David Robert Grimes, Andrea Hawkins-Daarud, VStefan Hoehme, Edwin F. Juarez, Carl Kesselman, Roeland Merks, Shannon M. Mumenthaler, Paul K. Newton, Kerri-Ann Norton, Rishi Rawat, Russell C. Rockne, Daniel Ruderman, Jacob Scott, Suzanne S. Sindi, Jessica L. Sparks, Kristin Swanson, David B. Agus, Paul Macklin
HI-C 2.0: AN OPTIMIZED HI-C PROCEDURE FOR HIGH-RESOLUTION GENOME-WIDE MAPPING OF CHROMOSOME CONFORMATION. Houda Belaghzal, Job Dekker, Johan H. Gibcus
The Image Data Resource: A Scalable Platform for Biological Image Data Access, Integration, and Dissemination. Eleanor Williams, Josh Moore, Simon W Li, Gabriella Rustici, Aleksandra Tarkowska, Anatole Chessel, Simone Leo, Balint Antal, Richard K Ferguson, Ugis Sarkans, Alvis Brazma, Rafael E Carazo-Salas, Jason Swedlow
SINCERITIES: Inferring gene regulatory networks from time-stamped single cell transcriptional expression profiles. Nan Papili Gao, Minhaz S.M. Ud-Dean, Rudiyanto Gunawan
Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Lindsey Montefiori, Liana Gonzales, Zijie Zhang, Yoav Gilad, Carole Ober, Gregory Crawford, Marcelo Nobrega, Noboru Jo Sakabe
SCODE: An efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Hirotaka Matsumoto, Hisanori Kiryu, Chikara Furusawa, Minoru S.H. Ko, Shigeru B.H. Ko, Norio Gouda, Tetsutaro Hayashi, Itoshi Nikaido
Scalable variational inference for super resolution microscopy. Ruoxi Sun, Evan Archer, Liam Paninski
Flowtrace: simple visualization of coherent structures in biological fluid flows. William Gilpin, Vivek N. Prakash, Manu Prakash
Tunable Extracellular Self-Assembly of Multi-Protein Conjugates from Bacillus subtilis. Charlie Gilbert, Mark Howarth, Colin Harwood, Tom Ellis
Multispot single-molecule FRET: high-throughput analysis of freely diffusing molecules. Antonino Ingargiola, Eitan Lerner, SangYoon Chung, Francesco Panzeri, Angelo Gulinatti, Ivan Rech, Massimo Ghioni, Shimon Weiss, Xavier Michalet
The Monarch Initiative: Insights across species reveal human disease mechanisms. Christopher Mungall,Julie McMurry, Sebastian Koehler, James Balhoff, Charles Borromeo, Matthew Brush, Seth Carbon, TOM CONLIN, Nathan Dunn, Mark Engelstad, Erin Foster, Jean-Philippe Gourdine, Julius Jacobsen, Daniel Keith, Bryan Laraway, Suzanna Lewis, Jeremy Nguyen Xuan, eKent Shefchek, Nicole Vasilevsky, Zhou Yuan, Nicole Washington, Harry Hochheiser,Tudor Groza, Damian Smedley, Peter Robinson, Melissa Haendel
Away from the bench
Ten simple rules for structuring papers. Konrad P Kording, Brett Mensh
Can paid reviews promote scientific quality and offer novel career perspectives for young scientists? Christian Wurzbacher, Hans-Peter Grossart, Erik Kristiansson, Henrik R Nilsson, Martin Unterseher
Starting from the end: what to do when restricted data is released. Marta Teperek, Rhys Morgan, Michelle Renee Ellefson,Danny Kingsley
Scientific data science and the case for Open Access. Gopal P Sarma
Why not…
How lizards fly: A novel type of wing in animals. J Maximilian Dehling


 (No Ratings Yet)
(No Ratings Yet)



 (1 votes)
(1 votes)













 It has long been considered that epithelial cells either migrate collectively as epithelial cells, or undergo an epithelial-to-mesenchymal transition and migrate as individual mesenchymal cells. Here,
It has long been considered that epithelial cells either migrate collectively as epithelial cells, or undergo an epithelial-to-mesenchymal transition and migrate as individual mesenchymal cells. Here,  A strong connection exists between the cell cycle and cell fate decisions in a wide-range of developmental contexts. Terminal differentiation is often associated with cell cycle exit, whereas cell fate switches are frequently linked to cell cycle transitions in dividing cells. In recent years, progress to address the connection between cell fate and the cell cycle has been made in pluripotent stem cells, in which the transition through mitosis and G1 phase is crucial for establishing a window of opportunity for pluripotency exit and the initiation of differentiation. Here,
A strong connection exists between the cell cycle and cell fate decisions in a wide-range of developmental contexts. Terminal differentiation is often associated with cell cycle exit, whereas cell fate switches are frequently linked to cell cycle transitions in dividing cells. In recent years, progress to address the connection between cell fate and the cell cycle has been made in pluripotent stem cells, in which the transition through mitosis and G1 phase is crucial for establishing a window of opportunity for pluripotency exit and the initiation of differentiation. Here, 






 (6 votes)
(6 votes)