PhD studentship investigating Somitogenesis on a Chip
Posted by Kim Dale, on 24 March 2014
Closing Date: 15 March 2021
Organisation: College of Life Sciences, University of Dundee
Supervisors: Dr Kim Dale and Dr Philip Murray
Studentship starting: 1st October 2014
The BBSRC East of Scotland Bioscience Doctoral Training Partnership provides training for postgraduates with diverse background (including biosciences, physical sciences, mathematics) to address biological questions using a range of technologies. This 4-year programme consists of a PhD project supplemented by Biosciences and Generic Skills Training, including a 3-month professional internship outside of academia
Application Deadline: 10th April 2014
Project
Oscillators are ubiquitous throughout biology (e.g. cardiac rhythms, circadian rhythms, the cell cycle). By definition they are dynamic and nonlinear in nature, making their behaviour nontrivial to quantify and understand.
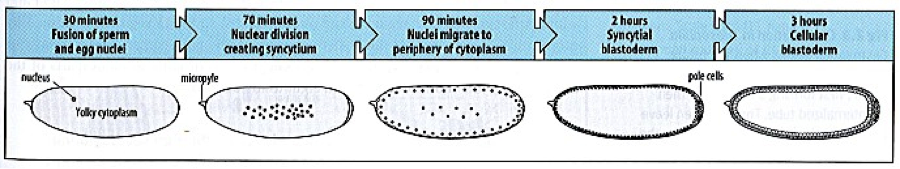
During vertebrate embryonic development, the formation of segmented blocks of mesodermal tissue (known as somites) occurs according to a strict temporal and spatial sequence and is regulated by a molecular oscillator known as the somitogenesis clock. The somites are transient structures that proceed to form essential segmented trunk tissues, such as the ribs and vertebrae of the skeleton, skeletal muscle, tendons and dermis. The process of somitogenesis is currently a field of high impact multidisciplinary research for a variety of reasons: for example, aberrations that arise during the segmentation process can give rise to medical conditions, such as scoliosis, in which the curvature of the spine is abnormal and while the etiology of many of these syndromes is largely unknown, linkage analyses have attributed some of these pathologies to mutations in key, highly conserved, segmentation clock genes; the system allows one to probe the fundamental questions of how heterogeneous spatial structure can emerge in an embryo and how the emergence of structure is coupled to embryo growth; the strict, regular, and cell co-ordinated spatio-temporal ordering with which somite formation occurs provides a unique means to quantitatively probe fundamental biological processes, such as gene transcription and mRNA processing, in in vivo contexts; modern observations demonstrate that a rich dynamical system, that is both amenable to and requiring of mathematical analysis, underlies the formation of morphological structure during somitogenesis.
It is now widely accepted that the spatio-temporal periodicity by which somites form is governed by oscillatory patterns of gene expression, regulated by a molecular oscillator known as the somitogenesis clock. Recent advances in the field have demonstrated that small groups of cells, taken from the most immature region of the pre-somitic tissue of a mouse embryo, undergo emergent patterning when cultured ex vivo in a plastic culture dish, a process that can be visualised in real time using genetically-modified reporter mice. These observations require the analysis of large datasets (i.e. real-time movies) and the use of mathematical models to interpret the spatio-temporal dynamics.
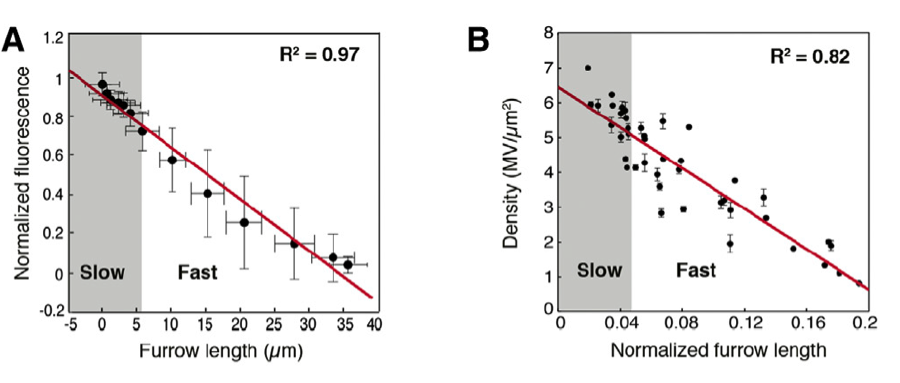
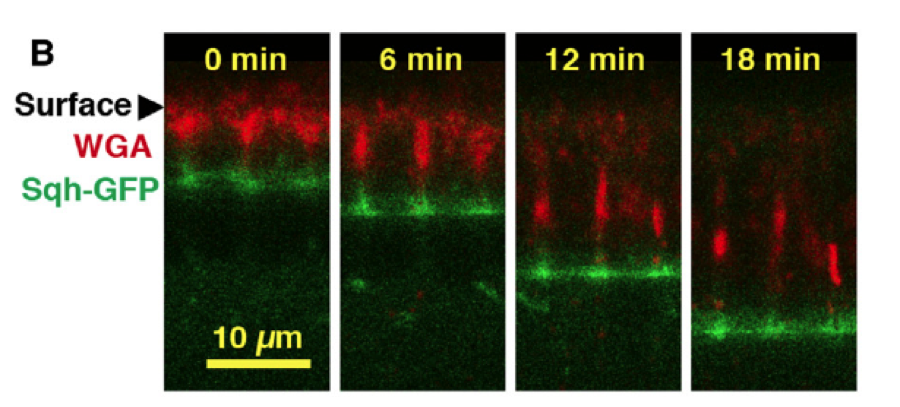
The goal of this study is to improve upon the current protocol by using lithographic techniques to fabricate a microfluidic channel that will mimic the 3D geometry in which the explanted tissue resides in vivo. This system will be used to house the tissue explant and will allow us to probe the underlying emergent behaviour that regulates somitogenesis in previously impossible ways. For example, recent work in the Dale lab has demonstrated that particular drugs modify the pace of the somitogenesis clock oscillations. Whilst these studies have been limited to snap shot views of the process, the developed technology will enable careful control of drug delivery and real-time monitoring of effect. There are numerous other means by which the developed toolkit will be used to probe fundamental questions regarding the emergence, propagation, degree of cell autonomy, and maintenance of somitogenesis clock oscillations.
The prospective student will benefit from respective expertise available at CLS (KD), Mathematics (PM) and Physics (DMG). KD will provide training in embryological techniques, PM will provide training in the use of computational software and mathematical modelling and DMG will provide training in the development and use of the microfluidic devices that will be used to house the explant. The student will benefit from being part of vibrant research groups in each of the individual disciplines and having access to a wide range of resources in the individual divisions
The project will generate high-quality, quantitative datasets that, together with mathematical modeling techniques, will enable us to measure and probe the somitogenesis clock oscillator. The developed techniques will allow us to integrate understanding of the molecular networks that generate oscillatory phenomena at the subcellular scale with observed emergent tissue-scale patterns. Using a predict-measure-refine workflow, the project will enable us to test and refine existing models of somitogenesis clock oscillations.
This project is ideal for a candidate with strong interests in cell/developmental Biology and the use of mathematical modelling to study biological questions in vivo. An enthusiasm for science and an enquiring mind is essential. No prior knowledge of chick or mouse development is required. This will involve a significant amount of imaging using confocal microscopy, alongside standard cell biological techniques such as whole mount immunostaining and in situ hybridisation. It will also involve image analysis and quantitation and modelling of the data sets.
Entry Requirements
Candidates must have a first or upper second class honours degree .
To apply
Interested candidates should in the first instance contact Kim Dale (j.k.dale@dundee.ac.uk).
For formal applications, visit:http://www.lifesci.dundee.ac.uk/phdprog/apply



 (No Ratings Yet)
(No Ratings Yet)
 (5 votes)
(5 votes)
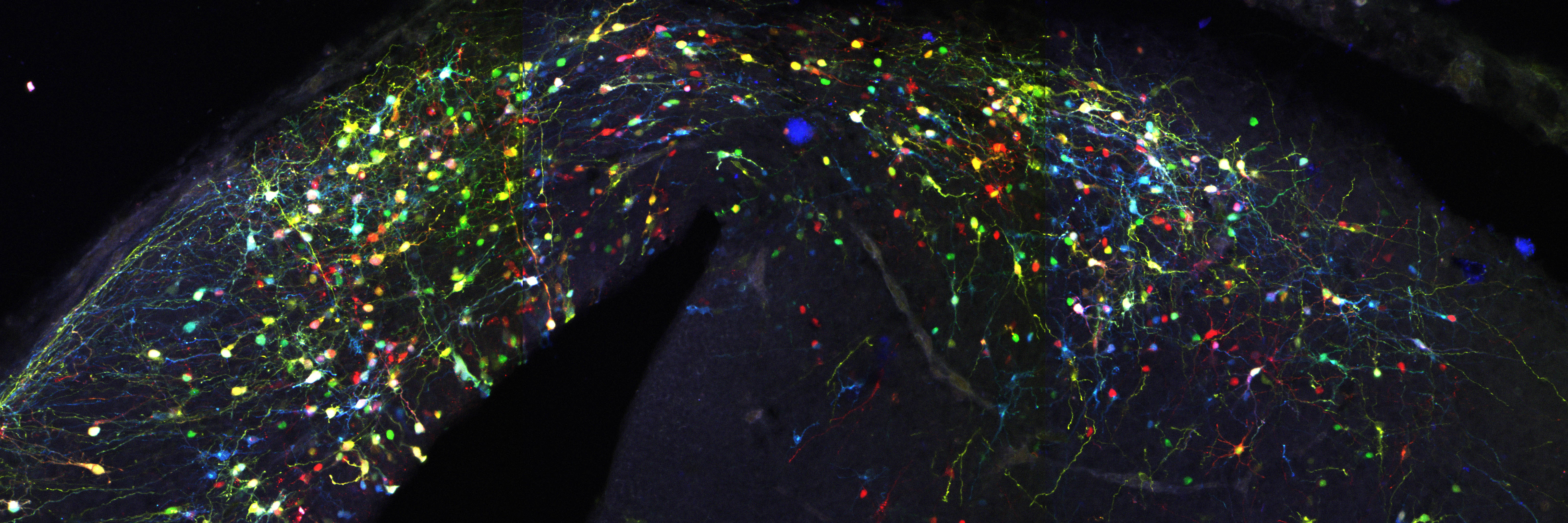
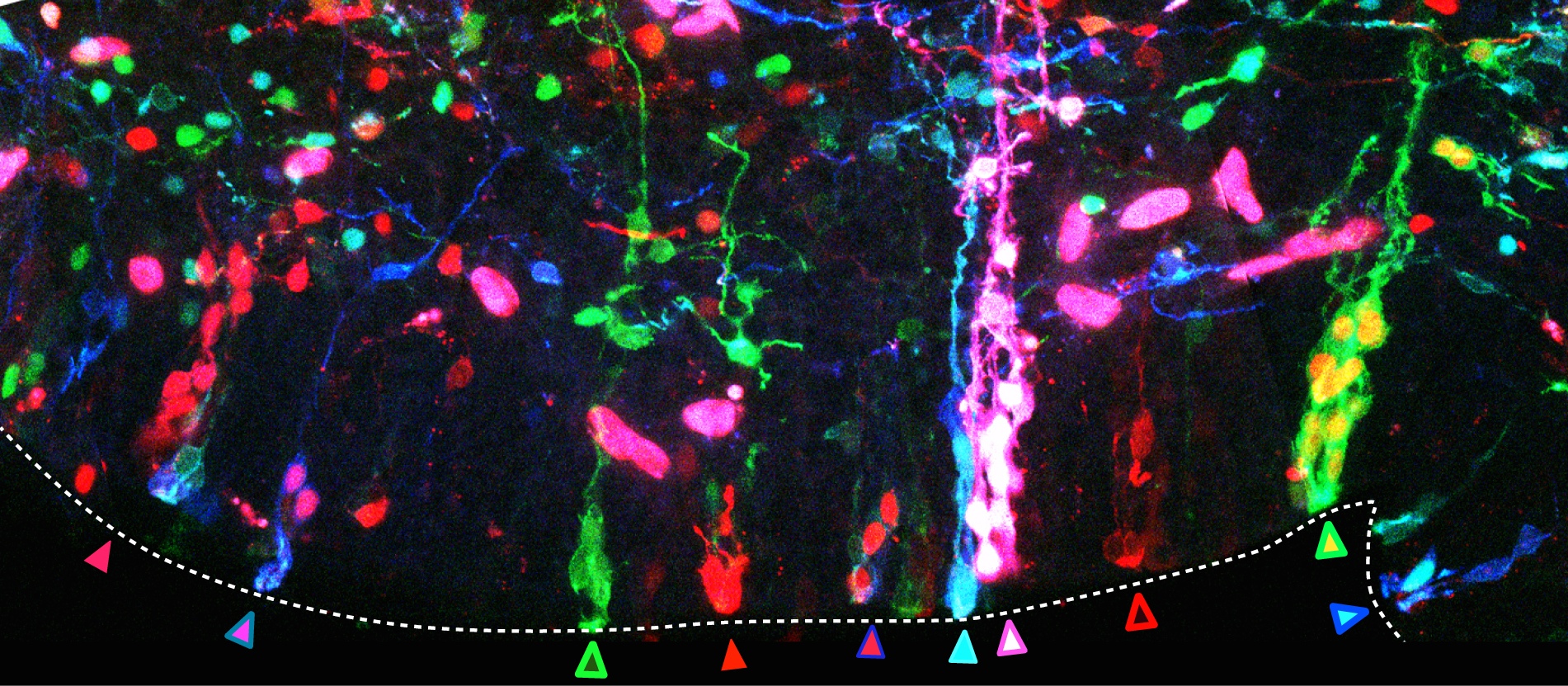
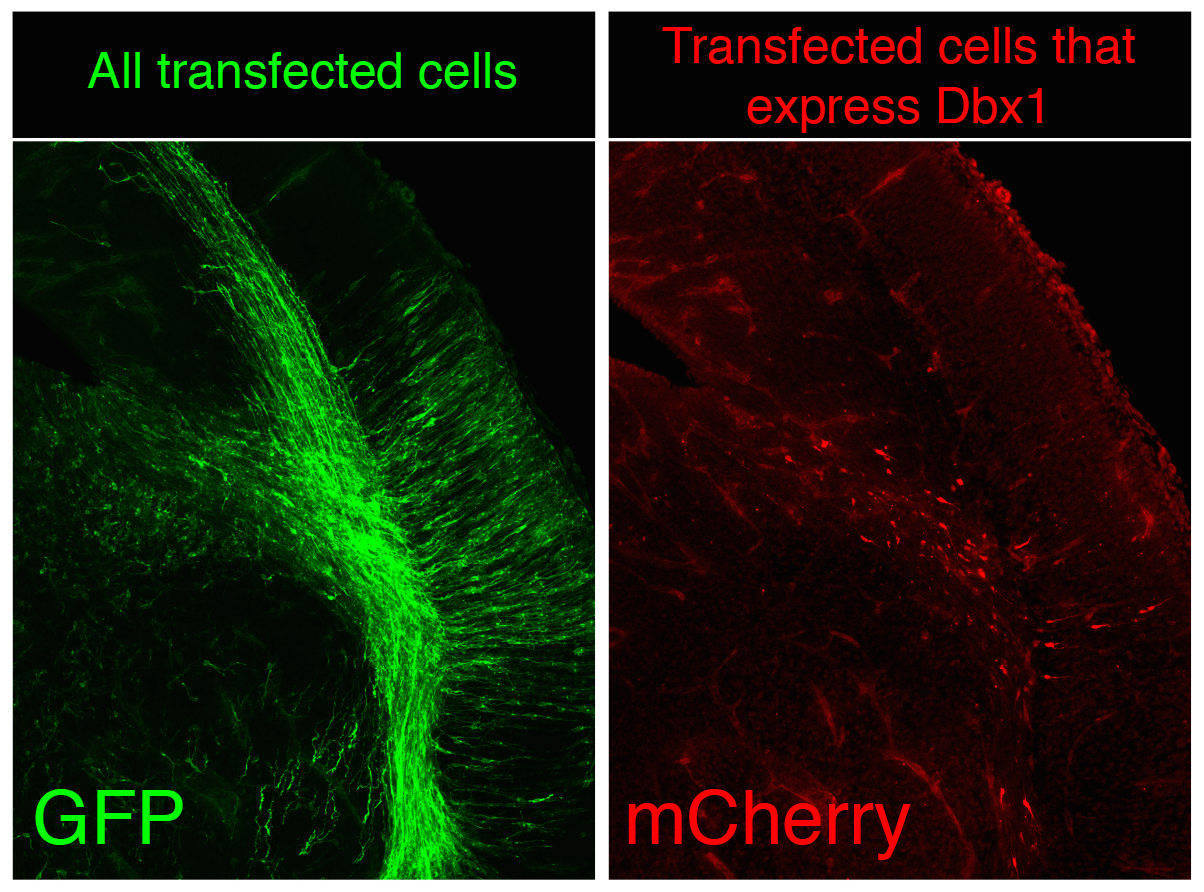
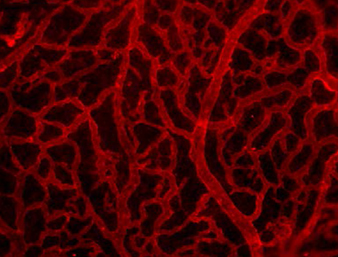
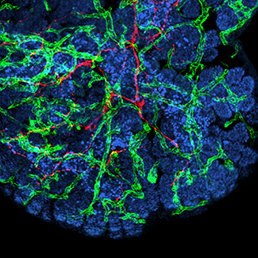 In many organs, the processes of vascularisation and innervation are frequently interdependent. In the pancreas, islet endocrine cells secrete vascular endocrine growth factor (VEGF) to direct vascularisation, but little is known about how islet innervation is regulated. On p.
In many organs, the processes of vascularisation and innervation are frequently interdependent. In the pancreas, islet endocrine cells secrete vascular endocrine growth factor (VEGF) to direct vascularisation, but little is known about how islet innervation is regulated. On p. 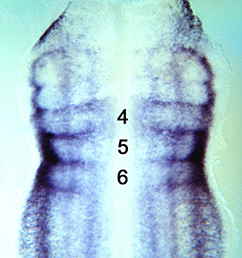
 Clathrin-mediated endocytosis regulates the signalling activity and turnover of multiple plasma membrane proteins. Interfering with endocytosis can therefore have complex effects on developmental processes. William Sessa and colleagues (p.
Clathrin-mediated endocytosis regulates the signalling activity and turnover of multiple plasma membrane proteins. Interfering with endocytosis can therefore have complex effects on developmental processes. William Sessa and colleagues (p. 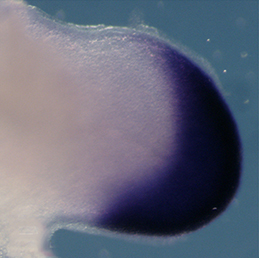 The vertebrate limb is an important model for understanding developmental patterning processes. Along the proximo-distal axis, the limb is segmented into stylopod (upper limb), zeugopod (lower limb) and autopod (hand/foot) regions, marked by the expression of particular homeobox genes – Meis1/2 most proximally and Hoxa13 most distally. Fibroblast growth factor (FGF) is a key inducer of distal fate, while the role of retinoic acid (RA) in promoting proximal fate has been controversial. Miguel Torres and co-workers (p.
The vertebrate limb is an important model for understanding developmental patterning processes. Along the proximo-distal axis, the limb is segmented into stylopod (upper limb), zeugopod (lower limb) and autopod (hand/foot) regions, marked by the expression of particular homeobox genes – Meis1/2 most proximally and Hoxa13 most distally. Fibroblast growth factor (FGF) is a key inducer of distal fate, while the role of retinoic acid (RA) in promoting proximal fate has been controversial. Miguel Torres and co-workers (p. 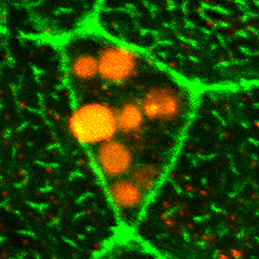 The Xenopus embryonic epidermis is a mucociliary epithelium – analogous to that found in mammalian airways. This tissue is characterised by the presence of multiciliated cells (MCCs) and goblet cells. A third cell type, the ion-secreting cell, was recently discovered in the Xenopus epidermis. Two papers now identify a final cell type in the frog embryonic skin, the small secretory cell (SSC). Eamon Dubaissi and colleagues (p.
The Xenopus embryonic epidermis is a mucociliary epithelium – analogous to that found in mammalian airways. This tissue is characterised by the presence of multiciliated cells (MCCs) and goblet cells. A third cell type, the ion-secreting cell, was recently discovered in the Xenopus epidermis. Two papers now identify a final cell type in the frog embryonic skin, the small secretory cell (SSC). Eamon Dubaissi and colleagues (p. 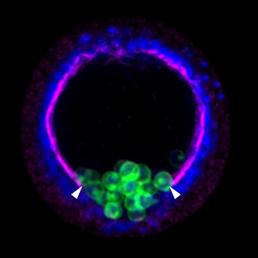 Epithelial-mesenchymal transition (EMT) is a cell state change used repeatedly during development across evolution, while aberrant EMT is associated with cancer metastasis. The transcriptional control of EMT has been extensively studied, and several transcription factors (TFs) shown to play crucial roles; notably, TFs such as Twist and Snail have been proposed to be ‘master regulators’ of EMT. On p.
Epithelial-mesenchymal transition (EMT) is a cell state change used repeatedly during development across evolution, while aberrant EMT is associated with cancer metastasis. The transcriptional control of EMT has been extensively studied, and several transcription factors (TFs) shown to play crucial roles; notably, TFs such as Twist and Snail have been proposed to be ‘master regulators’ of EMT. On p.  Haploid genetics holds great promise for understanding genome evolution and function. Much of the work on haploid genetics has previously been limited to microbes, but possibilities now extend to mammals. Here, Anton Wutz examines the potential use of haploid cells and puts them into a historical and biological context. See the Development at a Glance poster article on p.
Haploid genetics holds great promise for understanding genome evolution and function. Much of the work on haploid genetics has previously been limited to microbes, but possibilities now extend to mammals. Here, Anton Wutz examines the potential use of haploid cells and puts them into a historical and biological context. See the Development at a Glance poster article on p. 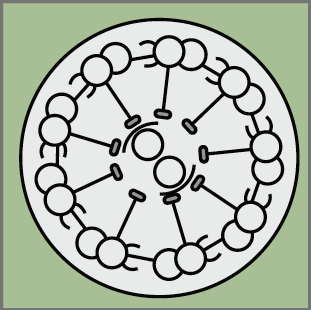 Cilia play many essential roles in fluid transport and cellular locomotion, and as sensory hubs for a variety of signal transduction pathways. Here, Sudipto Roy and colleagues review our understanding of the transcriptional control of ciliary biogenesis, highlighting the activities of FOXJ1 and the RFX family of transcriptional regulators. See the Review article on p.
Cilia play many essential roles in fluid transport and cellular locomotion, and as sensory hubs for a variety of signal transduction pathways. Here, Sudipto Roy and colleagues review our understanding of the transcriptional control of ciliary biogenesis, highlighting the activities of FOXJ1 and the RFX family of transcriptional regulators. See the Review article on p.