Towards a synthetic embryo
Posted by Aryeh Warmflash, on 24 September 2014
Waddington, whose writings on the epigenetic landscape continue to influence developmental biology to this day, called the developing embryo “the most intriguing object that nature has to offer”(Waddington, 1966). The mechanisms of pattern formation and morphogenesis have fascinated biologists for centuries. One question that is difficult to answer is what are the minimal requirements for generating an embryo? To ask a more specific question, at gastrulation stages, the cells of the mammalian embryo separate into three germ layers in a precise spatial arrangement. Is the information for this arrangement present in the cells themselves or do they need cues from the surrounding environment? With the ability to culture mammalian embryonic stem cells (ESCs), we can now begin to tackle these questions by asking how much of embryonic development we can recapitulate with ESCs in a culture dish.
Nearly every paper on ESCs touts their potential for regenerative medicine and there is a large literature that seeks to differentiate ESCs to pure populations of useful cells for just this purpose(e.g. Chambers et al., 2009; D’Amour et al., 2005; Kattman et al., 2011; Ozair et al., 2012). However, embryos, and by extension ESCs, are not in the business of making pure populations but have evolved to self-organized into patterns. Thus, given the right conditions, it is possible that embryonic patterns can be generated spontaneously in culture starting from ESCs. In a recent paper in Nature Methods(Warmflash et al., 2014), we asked whether the embryonic organization of the germ layers in the very early embryo can be achieved in a culture dish beginning with pluripotent human cells.
To begin to explore this idea, we began by simply adding BMP4, a part of the embryonic signaling cascade that induces gastrulation and patterning(Arnold and Robertson, 2009), to cultures of human ESCs grown under standard conditions. The cells readily differentiated, however, the results lacked reproducible organization at the level of fate. This negative result raised the question – why do ESCs fail to generate reproducible patterns in contrast to their embryonic analogs? One difference between the embryonic and culture environments is that the embryo provides a reproducible geometric structure while typical stem cell colonies take on arbitrary sizes and shapes (Figure 1A). We thus aimed to see how much of embryonic patterning could be restored simply by controlling the colony geometry (Figure 1B). To do so, we used micropatterning technology to grow cells on 1mm or smaller patches of cell adhesion molecules (Matrigel or Laminin) surrounded by a lawn of an inert substance to which cells and protein will not adhere. While such assays had been previous employed to evaluate the effects of varying colony size on cell fate(Bauwens et al., 2008), this work did not report any spatial structure in the differentiated colonies. To our astonishment, simple geometric confinement was sufficient to almost fully restore patterning. Two days after the cells were induced to differentiate with BMP4, four distinct cell populations emerged in concentric rings along the axis of the colony. From the outside in, these layers are extraembryonic, endoderm, mesoderm, and ectoderm (Figure 1C). This is precisely the arrangement found in the mammalian embryo at the conclusion of gastrulation.
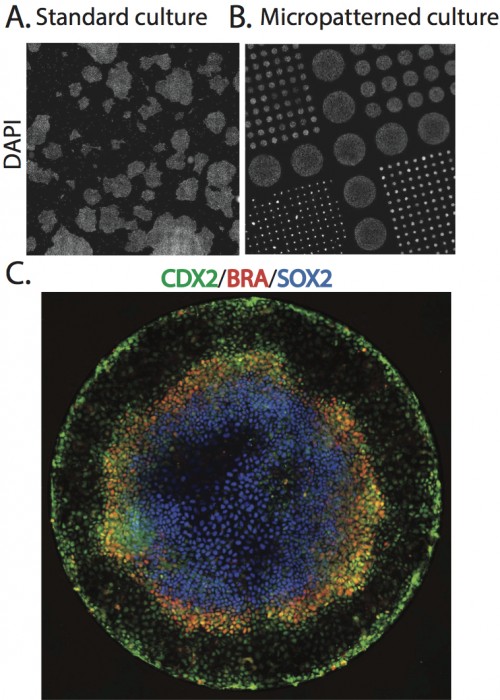
A. Scan of a large region of a standard hESC culture showing variable colony organization. B. Scan of a large region of micropatterned hESC culture. C. A single micro patterned colony (corresponding to the largest colonies in B) stained for markers of trophectoderm (Cdx2), mesoderm(BRA), and ectoderm (Sox2).
Since the differentiating signal is added at high levels to the medium, it is presented homogenously to all cells. Thus, the presence of four cell-fate territories implies that induced signaling between cells overwhelms exogenously supplied signals. How do cells use these induced signals to define position inside a colony 1 mm in size and containing approximately 2000 cells? A clue emerged from examining smaller colonies patterned onto the same slide. The smaller disks lost the inner-most fate, but preserved the outer three at the same position as measured from the colony edge! Thus, cell fates are defined using the colony border as a reference.
What signals are responsible for this long ranged communication? The direct response to BMP4 is measured by the nuclear accumulation of the activated transcription factor Smad1. Persistent BMP signaling is only seen in a narrow ring at the border of the colony and specifies the extraembryonic tissue found there. Inside the colony, a broader ring of activated Smad2, an upstream component of the Nodal signaling pathway, is responsible for inducing mesendoderm. The inner ectoderm territory is defined by the absence of both signals. Several decades of work in developmental genetics have revealed the identity of the inhibitors that restrict BMP and Nodal signaling in the embryo. When the combinations of inhibitors that give a robust embryonic phenotype(Bachiller et al., 2000; Perea-Gomez et al., 2002) are knocked down, cell fate markers spread in the expected way. Thus the boundary is defined by the loss of BMP inhibitors, while Nodal inhibitors restrict the range of the mesendodermal territory. This conclusion is reinforced by differentiating the colonies in mini-wells with high walls: when the inhibitors are prevented from escaping, the entire colony adopts the central ectodermal fate.
Beyond simply providing a reliable geometric structure, colony confinement also has profound effects on the fates that form. Unpatterned colonies form mesoderm and extraembryonic tissue (consistent with previous studies on BMP4 differentiation) but largely fail to form endodermal and ectodermal tissues. We speculate that the increased cell density that results from confinement is necessary both to achieve high enough levels of Nodal signaling to form the endodermal region, and high enough levels of inhibitors to establish the ectoderm.
But what is the relevance of a circularly symmetric colony to an embryo? The ring of mesodermal cells defined by Brachyury expression becomes mounded and displays a collection of markers such as Snail and activated Erk that are characteristic of the primitive streak (PS). Thus the micropatterned colonies appear to form a radially symmetric PS region. This is reminiscent of mutant mouse embryos that are unable to initiate the formation of an anterior-posterior (AP) axis and show a proximal and radially symmetric primitive streak(Nowotschin et al., 2013).
These experiments raise many questions. In the mouse embryo, the signals that initiate AP axis induction reside at the distal end of the visceral endoderm, the extraembryonic epithelial layer, and move to the future anterior side during gastrulation. Will the same signals, presented locally to a micropatterned colony, restrict the PS-streak like region and induce an axis? How does endogenous signaling compete with exogenously supplied BMP4 in the culture medium? The effects of secondary signals can be directly examined by growing cells an imposed flow with microfluidics to remove such signals(Moledina et al., 2012; Przybyla and Voldman, 2012). It is typically very hard to study how signals spread in intact embryos at endogenous levels, so synthetic systems permit one to test hypothesis and then return to the embryo with more targeted experiments.
Much of what we know about the mechanics and biochemistry of signaling comes from cell culture experiments, which have at best only a temporal dimension. But the spatial patterns formed by morphogens and their secondary inhibitors are a biologically relevant phenotype, for which a simple in vitro assay has been lacking until now. Our system is very amenable to long term time lapse imaging, and it will be possible follow markers for cell fates as they emerge in time, and ask, for instance, whether the speckled patterns seen with immunofluorescence are merely cells following the same trajectories but at different rates. It is also an ideal system in which to quantify signaling in space and time and thus in a more predictive way engineer tissues and organs.
Arnold, S. J. and Robertson, E. J. (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10, 91–103.
Bachiller, D., Klingensmith, J., Kemp, C., Belo, J. A., Anderson, R. M., May, S. R., McMahon, J. A., McMahon, A. P., Harland, R. M., Rossant, J., et al. (2000). The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature 403, 658–661.
Bauwens, C. L., Peerani, R., Niebruegge, S., Woodhouse, K. A., Kumacheva, E., Husain, M. and Zandstra, P. W. (2008). Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells 26, 2300–2310.
Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M. and Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280.
D’Amour, K. A., Agulnick, A. D., Eliazer, S., Kelly, O. G., Kroon, E. and Baetge, E. E. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23, 1534–1541.
Kattman, S. J., Witty, A. D., Gagliardi, M., Dubois, N. C., Niapour, M., Hotta, A., Ellis, J. and Keller, G. (2011). Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240.
Moledina, F., Clarke, G., Oskooei, A., Onishi, K., Günther, A. and Zandstra, P. W. (2012). Predictive microfluidic control of regulatory ligand trajectories in individual pluripotent cells. Proc Natl Acad Sci USA 109, 3264–3269.
Nowotschin, S., Costello, I., Piliszek, A., Kwon, G. S., Mao, C.-A., Klein, W. H., Robertson, E. J. and Hadjantonakis, A.-K. (2013). The T-box transcription factor Eomesodermin is essential for AVE induction in the mouse embryo. Genes Dev 27, 997–1002.
Ozair, M. Z., Noggle, S., Warmflash, A., Krzyspiak, J. E. and Brivanlou, A. H. (2012). SMAD7 Directly Converts Human Embryonic Stem Cells to Telencephalic Fate by a Default Mechanism. Stem Cells 31, 35–47.
Perea-Gomez, A., Vella, F. D. J., Shawlot, W., Oulad-Abdelghani, M., Chazaud, C., Meno, C., Pfister, V., Chen, L., Robertson, E., Hamada, H., et al. (2002). Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell 3, 745–756.
Przybyla, L. M. and Voldman, J. (2012). Attenuation of extrinsic signaling reveals the importance of matrix remodeling on maintenance of embryonic stem cell self-renewal. Proc Natl Acad Sci USA 109, 835–840.
Waddington, C. H. (1966). Principles of Development and Differentiation. New York: Macmillan.
Warmflash, A., Sorre, B., Etoc, F., Siggia, E., & Brivanlou, A. (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells Nature Methods, 11 (8), 847-854 DOI: 10.1038/nmeth.3016


 (7 votes)
(7 votes)
 (3 votes)
(3 votes)
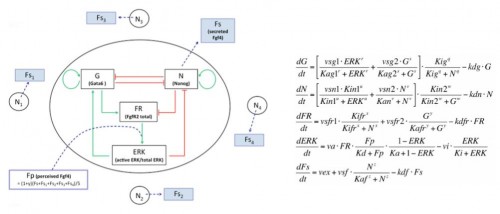
 In the early embryo, the first fate decision separates the trophectoderm from the inner cell mass (ICM). Subsequently, the ICM segregates into epiblast (Epi) and primitive endoderm (PrE), but how do cells decide which of these two fates to adopt? Claire Chazaud, Geneviève Dupont and co-workers address this question in the early mouse embryo (see p.
In the early embryo, the first fate decision separates the trophectoderm from the inner cell mass (ICM). Subsequently, the ICM segregates into epiblast (Epi) and primitive endoderm (PrE), but how do cells decide which of these two fates to adopt? Claire Chazaud, Geneviève Dupont and co-workers address this question in the early mouse embryo (see p.  Intra-cytoplasmic sperm injection (ICSI) is a method of fertilisation that involves the injection of a single sperm into an oocyte. The technique has been applied successfully to generate viable offspring in human, mouse and other vertebrates, but never before has a live bird been produced using ICSI due to difficulties in mimicking avian polyspermic fertilisation. Now, on p.
Intra-cytoplasmic sperm injection (ICSI) is a method of fertilisation that involves the injection of a single sperm into an oocyte. The technique has been applied successfully to generate viable offspring in human, mouse and other vertebrates, but never before has a live bird been produced using ICSI due to difficulties in mimicking avian polyspermic fertilisation. Now, on p. Physical properties such as tissue stiffness have been shown to influence stem cell fate in vitro. It is possible that spatiotemporal changes in stiffness may influence tissue formation during development; however, it has been difficult to analyse this due to the technical limitations of measuring stiffness in embryonic tissues. In this issue (p.
Physical properties such as tissue stiffness have been shown to influence stem cell fate in vitro. It is possible that spatiotemporal changes in stiffness may influence tissue formation during development; however, it has been difficult to analyse this due to the technical limitations of measuring stiffness in embryonic tissues. In this issue (p.  Gastrulation occurs when a single-layered blastula transitions to a gastrula containing all three germ layers: ectoderm, endoderm and mesoderm. Many aspects of vertebrate gastrulation have been well-characterised using the model vertebrate Xenopus, but surprisingly little is known about how the dorsal mesoderm moves from the outer surface to the interior of the gastrula. Now, on p.
Gastrulation occurs when a single-layered blastula transitions to a gastrula containing all three germ layers: ectoderm, endoderm and mesoderm. Many aspects of vertebrate gastrulation have been well-characterised using the model vertebrate Xenopus, but surprisingly little is known about how the dorsal mesoderm moves from the outer surface to the interior of the gastrula. Now, on p.  The mitotic-to-meiotic transition during spermatogenesis is essential for generating haploid spermatid cells from diploid spermatogonial cells. After differentiation and proliferation of spermatogonial cells, two rounds of meiosis are required to generate spermatid cells, but how the complex transition from mitosis to meiosis is coordinated remains unclear. In this issue (p.
The mitotic-to-meiotic transition during spermatogenesis is essential for generating haploid spermatid cells from diploid spermatogonial cells. After differentiation and proliferation of spermatogonial cells, two rounds of meiosis are required to generate spermatid cells, but how the complex transition from mitosis to meiosis is coordinated remains unclear. In this issue (p.  In May this year, Stockholm hosted a Keystone Symposium on Adult Neurogenesis, attracting scientists from around the world despite the lack of customary snow. The symposium offered an extraordinary program, covering diverse topics that ranged from the neural stem cell lineage and regulation of neurogenesis to functional aspects of neurogenesis in homeostasis and disease, and even computational modeling. Here
In May this year, Stockholm hosted a Keystone Symposium on Adult Neurogenesis, attracting scientists from around the world despite the lack of customary snow. The symposium offered an extraordinary program, covering diverse topics that ranged from the neural stem cell lineage and regulation of neurogenesis to functional aspects of neurogenesis in homeostasis and disease, and even computational modeling. Here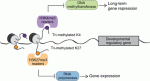 Poised (bivalent) chromatin is defined by the simultaneous presence of histone modifications associated with both gene activation and repression. This epigenetic feature was first observed at promoters of lineage-specific regulatory genes in embryonic stem cells in culture. More recent work has shown that,
Poised (bivalent) chromatin is defined by the simultaneous presence of histone modifications associated with both gene activation and repression. This epigenetic feature was first observed at promoters of lineage-specific regulatory genes in embryonic stem cells in culture. More recent work has shown that, 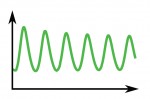 Biological clocks play key roles in organismal development, homeostasis and function. In recent years, much work has focused on circadian clocks, but emerging studies have highlighted the existence of ultradian oscillators – those with a much shorter periodicity than 24 h. Accumulating evidence, together with recently developed optogenetic approaches, suggests that such ultradian oscillators play important roles during cell fate decisions, and analyzing the functional links between ultradian oscillation and cell fate determination will contribute to a deeper understanding of the design principle of developing embryos. Here, Akihiro Isomura and
Biological clocks play key roles in organismal development, homeostasis and function. In recent years, much work has focused on circadian clocks, but emerging studies have highlighted the existence of ultradian oscillators – those with a much shorter periodicity than 24 h. Accumulating evidence, together with recently developed optogenetic approaches, suggests that such ultradian oscillators play important roles during cell fate decisions, and analyzing the functional links between ultradian oscillation and cell fate determination will contribute to a deeper understanding of the design principle of developing embryos. Here, Akihiro Isomura and (No Ratings Yet)
(No Ratings Yet)



