At the last BSDB meeting Adi was the winner of the BSDB poster prize. Unfortunately John Young – the winner of the last SDB poster prize – was unable to interview him, so The Node interviewed Adi on this occasion.
The Node: In which lab do you work and how long have you been there?
Adi: I am with Prof. Helen Skaer, at the department of Zoology in Cambridge. I’ve been there for six years now.
The Node: What does your lab work on?
Adi: We are largely interested in morphogenesis and cell fate decisions in developing organs, and we use Drosophila Malpighian (renal) tubules as a model. Tubules are special in many ways. They are an excellent model to study branching morphogenesis, re-structuring of epithelial tubes (which is what I do), apico-basal polarity during tubulogenesis and the relationship between organ structure and function. These are largely the areas that we address.
 The Node: What was your poster about and what was the focus of your PhD project?
The Node: What was your poster about and what was the focus of your PhD project?
Adi: I am studying the later development of Malpighian tubules. Early in development cells in tubule primordia divide to result in short thick tubes. In absence of any significant changes in cell number, cell rearrangements elongate these tubes to four times their original length. This process has been quite elusive at the cellular level. For my thesis I have been interested in understanding how you go from a short thick tube to a long thin one in the absence of cell division.
The Node: So the cell numbers don’t change?
Adi: No, they largely don’t change. There are some cells which come from the outside and integrate, but largely the tubule cells that exist in a 11 hour old embryo are the cells that will undergo rearrangements to give rise to a long tube later. I’ve been live imaging tubule elongation, which takes something like 5 to 6 hours, and I have imaged and tracked many cells to understand their behavior. It turns out that the tubule cells undergo convergent-extension movements, where cells intercalate between their neighbours to result in elongation.
The Node: It sounds like your PhD had a lot of microscopy. Did you have to develop any particular techniques, or did you have to use a specific type of microscope to be able to do this long-term imaging?
Adi: I tried working with microscopes that people think are good for imaging inside the body, like two-photon. But unfortunately for me nothing worked when I tried fancy microscopes. So I relied heavily on genetics. Because tubules are quite deep within the embryo, they are difficult to image. So I started making several recombinant lines using a tubule-specific GAL4 and various UAS-fluorescent reporters until I got a recombinant that showed very high expression. At this expression level, I did not need a fancy microscope, so I did most of my imaging on a confocal microscope. I think Drosophila genetics are very powerful. You can solve most of your problems by carefully thinking about how to use genetic tricks.
The Node: Had you actually won a prize at a meeting before?
Adi: Not something as big as the BSDB, but I have won prizes in college, back in India, when I was studying for my Bachelors. This is the biggest prize I have ever got. I am very pleased about it.
The Node: Have you completed this story yet?
Adi: We are hoping that a paper will be submitted very soon. But I should say that an important aspect of my work ties with the work of Dr. Barry Denholm, an independent researcher in Zoology associated with Helen Skaer’s lab. Moving cells in an elongating tissue need to know not only their top and bottom (apical-basal), but also their right and left (planar information). This is not understood very well in epithelial tubes. It is not known, for example, if all epithelial tubes are planar polarized. Barry could show that Malpighian tubule cells are planar polarized: they know their right from their left. He also figured out the signaling mechanism that is important for this planar polarization. Barry’s work gave us an important clue on what might regulate convergent-extension movements during tubule elongation.
The Node: Now that you are finishing your PhD and finishing your paper, what is your next plan?
Adi: I would like to continue doing research. I know a little bit about how development works, but to me it is not very clear yet how what we know from developmental systems relates to human health. For example, could the things we study in developmental biology explain how metastasis works? That excites me immensely- how you maintain a normal state, and how that state goes wrong to result in a disease. During development you have to deploy, re-deploy and shut down pathways all the time. It cannot be that the basic principles are very different in disease states. So, I would like to study the balance between disease & homeostasis, and how developmental processes are involved there.
The Node: And are you planning on staying here in the UK or you are thinking of going back to India or moving elsewhere…
Adi: During my thesis I spent around 2 years in India, in the lab of Professor VijayRaghavan, who is a developmental biologist working at NCBS in Bangalore. The most important part of my data came from working in Vijay’s lab. I really enjoyed my time there and I would very much like to spend at least a part of my postdoc period in India.
The Node: I noticed that you won a scholarship for Indian citizens to study in Cambridge. Was it very hard to get this scholarship?
Adi: I got a Nehru-Cambridge scholarship, which is awarded by the Cambridge Commonwealth Trust. This scholarship is given to very few people, but I think I got lucky! The Commonwealth Trust is incredible and the Nehru-Cambridge has been very supportive, especially during the troubled periods of my PhD. When I needed an extension they actually gave me generous financial support.
The Node: It seems that you had a good support network: in your lab, the people who financed your PhD…
Adi: Yes, and I think it is important because not everyone knows what is the right thing to do in the first instance. You begin by exploring a problem that you think matters. You are very ambitious when you start because you are also slightly naïve about what can be done. But then it turns out that some techniques are just too difficult to answer your question. That was the case with me- I started doing Mass Spectrometry, which I had never done before, and it was quite tough to adapt it for Drosophila embryonic Mapighian tubules. But I think it is important that you do what you think is right and if you fail hopefully people will realize that it was worth trying and still support you.
The Node: You won the BSDB poster prize, but there was another student in the Skaer lab that won the Beddington medal for best PhD thesis. What do you think is making this year such a successful year for your lab? Is there something in the water, or the cakes you guys eat here?!
Adi: I can tell you why Helen Weavers won the Beddington medal. She is one of the most focused and skillful colleagues I have ever worked with. I think she is very talented, both in terms of thinking about experiments and doing them. It was a very well thought through project, and I think that is why it was such a successful endeavor.
Success of the lab is certainly down to my boss Helen Skaer. Helen is a tremendously good supervisor who creates a very interactive environment in the lab. She is constructively critical about experiments and results, skillful at suggesting the next steps, resourceful at initiating collaborations and makes you feel good about what you are doing. She allows people to explore, she knows their abilities, and suggests the right things to them. If anything, my prize certainly belongs to her more than to me.
The Node: You will be attending the ISDB meeting in June. What are you expecting? You talk about the BSDB as being quite a big meeting, but this should be at a different scale.
Adi: Yes, absolutely. I have attended one ISDB meeting before, in Edinburgh. It changed my perspective, because it had sessions where people were talking about all sorts of things from miRNAs to evo-devo. I have checked the programme for this ISDB and it looks really exciting. Lots of people who will be there are my heroes, and I am really looking forward to hearing talks beyond epithelial morphogenesis.
At the ISDB meeting Adi will, in turn, interview the winner of the SDB poster prize.
 (7 votes)
(7 votes)
 Loading...
Loading...


 (4 votes)
(4 votes)
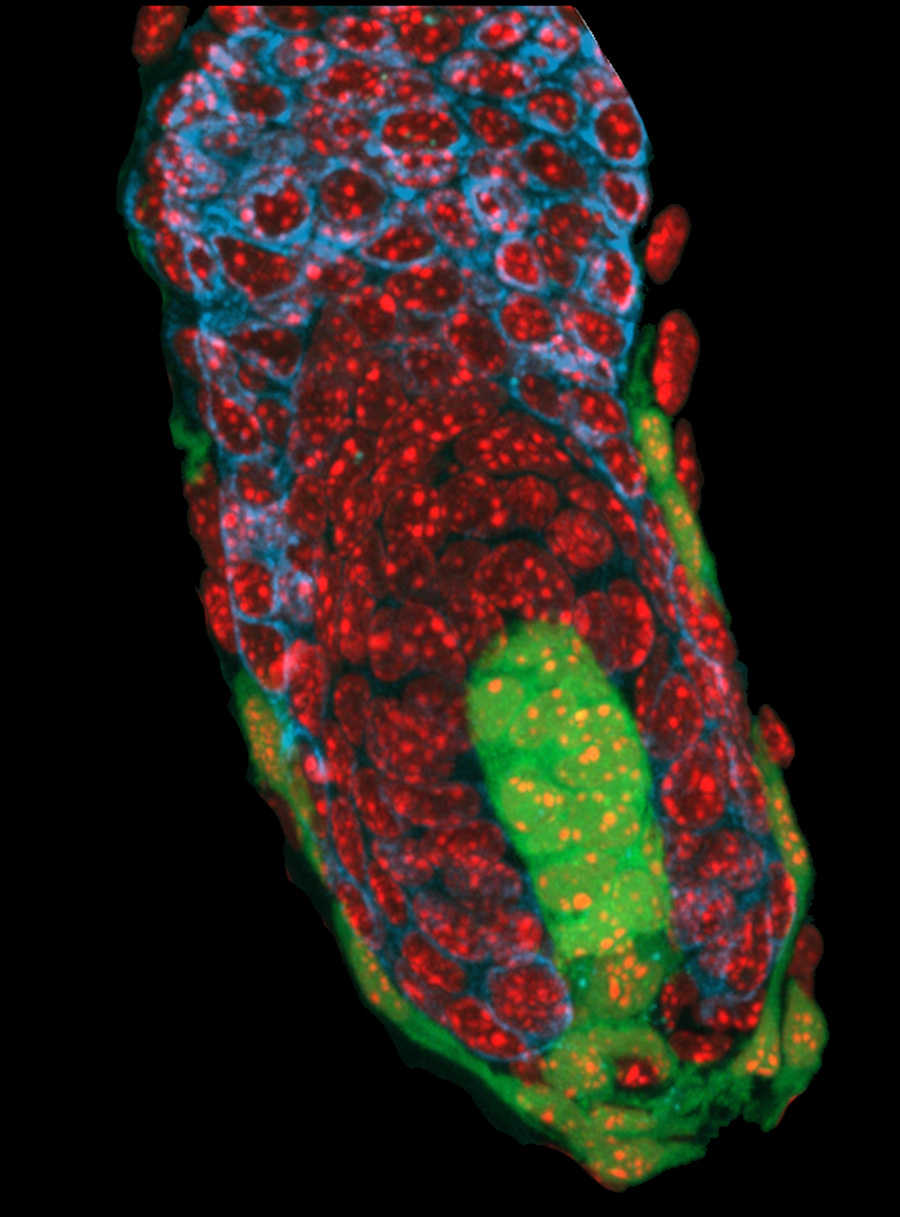
 Asymmetric cell divisions (ACDs) play a crucial role in controlling cell fate and generating cell diversity during development. The centrosome is known to be involved in ACD, and recent studies have shown that centrosomes exhibit dynamic and asymmetric movements that regulate orientation of the mitotic spindle. Here, Yohanns Bellaiche and co-workers identify a novel type of centrosome movement during cytokinesis (
Asymmetric cell divisions (ACDs) play a crucial role in controlling cell fate and generating cell diversity during development. The centrosome is known to be involved in ACD, and recent studies have shown that centrosomes exhibit dynamic and asymmetric movements that regulate orientation of the mitotic spindle. Here, Yohanns Bellaiche and co-workers identify a novel type of centrosome movement during cytokinesis ( Early embryonic development occurs in the absence of transcription; instead, it relies on maternal mRNAs and proteins present within the egg. It is believed that this period of transcriptional quiescence is maintained by factors that eventually become titrated out during early cleavages, thus leading to zygotic genome activation. How exactly this transition occurs, however, is unclear. Here, Jim Smith, Steven Harvey and co-workers use exome sequencing and RNA-seq to distinguish between maternal and zygotic transcriptomes in early zebrafish embryos (
Early embryonic development occurs in the absence of transcription; instead, it relies on maternal mRNAs and proteins present within the egg. It is believed that this period of transcriptional quiescence is maintained by factors that eventually become titrated out during early cleavages, thus leading to zygotic genome activation. How exactly this transition occurs, however, is unclear. Here, Jim Smith, Steven Harvey and co-workers use exome sequencing and RNA-seq to distinguish between maternal and zygotic transcriptomes in early zebrafish embryos ( Neural stem/progenitor cells in the mammalian hippocampus generate new neurons throughout life. But how do these integrate into a mature and functional neural circuitry? Here, Sebastian Jessberger and colleagues address this question by using a new imaging approach to analyse neurite growth from newborn granular cells (
Neural stem/progenitor cells in the mammalian hippocampus generate new neurons throughout life. But how do these integrate into a mature and functional neural circuitry? Here, Sebastian Jessberger and colleagues address this question by using a new imaging approach to analyse neurite growth from newborn granular cells (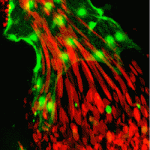 The correct formation of blood vessels is essential for the development of a functional vasculature. Various mechanisms of vascular lumen formation have been described to date but, now, Wiebke Herzog and colleagues examine the development of common cardinal veins (CCVs) in zebrafish and show that these form via a previously undescribed mode of lumen formation (
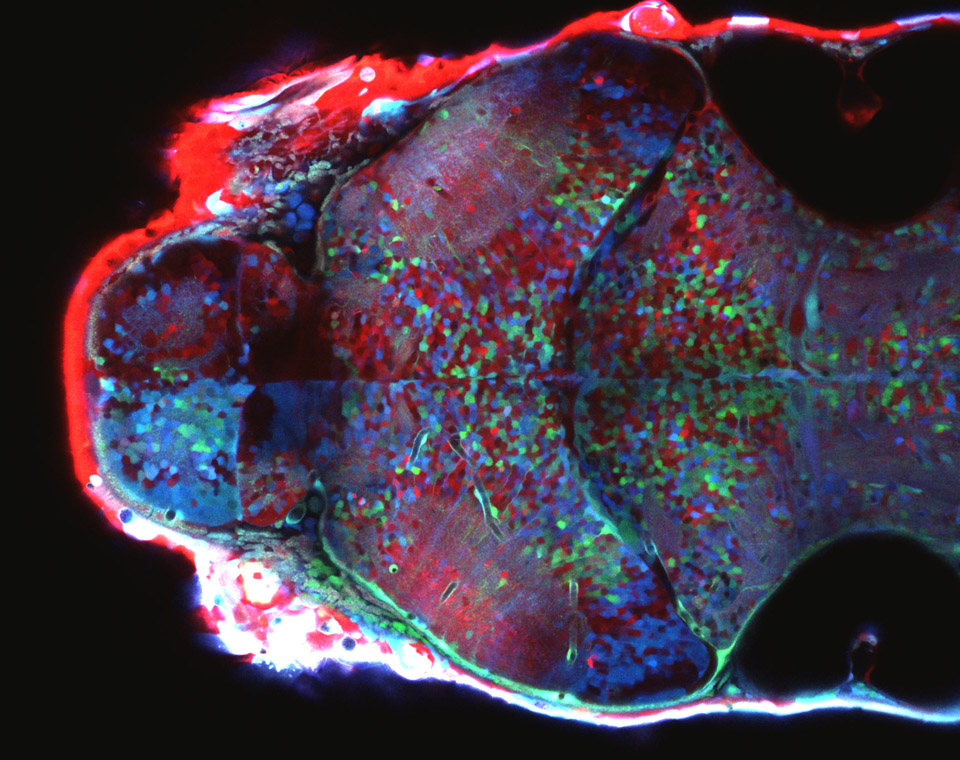
The correct formation of blood vessels is essential for the development of a functional vasculature. Various mechanisms of vascular lumen formation have been described to date but, now, Wiebke Herzog and colleagues examine the development of common cardinal veins (CCVs) in zebrafish and show that these form via a previously undescribed mode of lumen formation (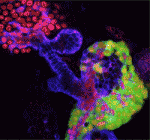 The liver and ventral pancreas are thought to develop from a common pool of multipotent progenitors. Although a number of studies have identified factors required for either pancreas or liver specification, factors that are distinctly required to specify the entire hepatopancreas system have not yet been reported. Now, Joseph Lancman and co-workers uncover a common genetic program, involving hnf1ba and wnt2bb, that specifies progenitors of the liver, ventral pancreas, gall bladder and associated ducts in zebrafish (
The liver and ventral pancreas are thought to develop from a common pool of multipotent progenitors. Although a number of studies have identified factors required for either pancreas or liver specification, factors that are distinctly required to specify the entire hepatopancreas system have not yet been reported. Now, Joseph Lancman and co-workers uncover a common genetic program, involving hnf1ba and wnt2bb, that specifies progenitors of the liver, ventral pancreas, gall bladder and associated ducts in zebrafish (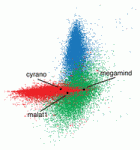 Long non-coding RNAs (lncRNAs) have recently emerged as key regulators of gene expression in embryos and in embryonic stem cells (ESCs). Recent large-scale genomics approaches have identified thousands of putative lncRNAs but are these all truly non-coding RNAs? Here, on
Long non-coding RNAs (lncRNAs) have recently emerged as key regulators of gene expression in embryos and in embryonic stem cells (ESCs). Recent large-scale genomics approaches have identified thousands of putative lncRNAs but are these all truly non-coding RNAs? Here, on 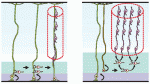 Song-Hai Shi and colleagues review recent findings on the generation, migration and organization of excitatory and inhibitory neurons in the neocortex, and discuss how the lineage history of neurons influences the assembly of functional circuits.
Song-Hai Shi and colleagues review recent findings on the generation, migration and organization of excitatory and inhibitory neurons in the neocortex, and discuss how the lineage history of neurons influences the assembly of functional circuits. Read the Editorial by our Editor-in-Chief, Olivier Pourquié on p.
Read the Editorial by our Editor-in-Chief, Olivier Pourquié on p. (No Ratings Yet)
(No Ratings Yet)