Here are the highlights from the current issue of Development:
Chewing the Fat over PCP and growth
The Drosophila protocadherin Fat (Ft) affects planar cell polarity (PCP) but also inhibits the overgrowth of imaginal discs by activating the Hippo pathway. The intracellular domain (ICD) mediates much of Ft activity, but are its PCP and Hippo pathway activities separable? To answer this question, Hitoshi Matakatsu and Seth Blair have undertaken a detailed structure-function analysis of the Ft ICD (see p. 1498). They identify PCP and Hippo pathway-specific domains in the Ft ICD and, surprisingly, report that these domains do not map to any previously identified protein interaction domains in the Ft ICD, nor, with one exception, to the regions that are highly conserved in mammalian Fat4. They also confirm and extend evidence showing that the extracellular domain of Ft has activities in both the PCP and Hippo pathways that are mediated by the protocadherin Dachsous. Based on their results, the authors conclude that the PCP and Hippo pathway activities of Fat and Dachsous are largely separable.
Robo-Slit signals regulate CNS motoneuron axon exit
In mammals, signals transmitted from the central nervous system (CNS) to muscles via motoneurons control movement. To form these circuits, motoneurons extend their axons out of the CNS at specialised exit points. Here (p. 1435), Zaven Kaprielian and colleagues use mouse spinal accessory motoneurons (SACMNs) to investigate how this essential phase of motor axon pathfinding is controlled. SACMNs, which innervate neck and back muscles, leave the spinal cord at lateral exit points (LEPs). In mice lacking the homeodomain transcription factor Nkx2.9, the researchers report, SACMN axons project normally to the LEP but fail to exit the CNS. Robo2 expression in SACMNs is downregulated in Nkx2.9 null mice, they report, and SACMN axons fail to exit the spinal cord in Robo2-deficient animals. Finally, the Robo2 ligands Slit1-3 are present at the LEP and SACMN axons fail to exit the CNS in Slit-deficient mice. Together, these results suggest that Nkx2.9 controls SACMN axon exit from the CNS by regulating Robo2-Slit signalling.
sRNA paths to plant female gamete development
During the first phase of Arabidopsis female gamete formation (megasporogenesis), a somatic ovule cell differentiates into a megaspore mother cell and divides to generate four haploid megaspores. In the next phase (megagametogenesis), one of these megaspores undergoes syncytial mitosis and differentiates to form the female gametophyte. It’s known that a somatic small RNA (sRNA) pathway restricts reproductive potential to this functional megaspore but what controls the megasporogenesis to megagametogenesis transition? Here (p. 1399), Matthew Tucker and co-workers examine gene expression patterns in ovule tissues and show that an sRNA pathway is also involved in this phase of female gamete formation. The researchers report that ARGONAUTE5 (AGO5), a putative sRNA pathway effector, is expressed around reproductive cells during megasporogenesis and show that a unique semi-dominant ago5-4 insertion allele disrupts the initiation of megagametogenesis. Expression of a viral RNAi suppressor protein in the somatic cells flanking the megaspores produces a similar phenotype. Thus, the researchers conclude, at least two somatic sRNA pathways contribute to female gametophyte development in Arabidopsis.
PHD-fingers point to meristem initiation
Apical meristems, which are indispensable for plant growth and development, contain stem cells and the organizer, both of which are specified during early embryogenesis. During root meristem initiation, specification of hypophysis (the precursor of the organizer) requires the transcription factor MONOPTEROS (MP), which functions in part by activating the expression of TARGET of MP (TMO) transcription factors. But how does MP activate the expression of these genes in the context of chromatin to regulate meristem initiation? On p. 1391, Yoshibumi Komeda and colleagues report that the plant homeodomain (PHD)-finger proteins OBERON (OBE) and TITANIA (TTA) are essential for MP-dependent embryonic root meristem initiation in Arabidopsis. They show that these PHD-finger proteins interact with each other and that, although MP expression is unaltered by mutations in OBE/TTA genes, the expression of TMO5 and TMO7 is locally lost in obe1 obe2 embryos. These and other data indicate that PHD-finger protein complexes control the activation of transcription factor genes during root meristem initiation.
Joining forces in the neural tube
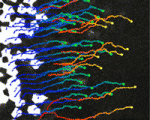
In developing vertebrates, neural tube closure (NTC) – the formation of the neural tube from a sheet of neural ectoderm – requires both neural ectoderm and non-neural ectoderm. But, whereas cell shape changes, cell rearrangement and cell division in the neural ectoderm are essential for NTC, the cellular changes in the non-neural ectoderm that contribute to NTC are unclear. Now, on p. 1417, Naoto Ueno and co-workers use digital scanned laser light sheet fluorescence microscopy to examine the movements of non-neural ectoderm cells in Xenopus embryos during NTC. The researchers show that the collective movement of deep-layer non-neural ectoderm cells towards the dorsal midline helps to drag along the superficial non-neural ectoderm during NTC. Inhibition of this movement by deletion of integrin β1 function, they report, blocks NTC completion. By contrast, oriented cell division, cell shape changes and cell rearrangement in the non-neural ectoderm have little or no role in NTC. Together, these results suggest that a global reorganisation of embryonic tissues is involved in NTC.
Neurula rotation breaks ascidian LR symmetry
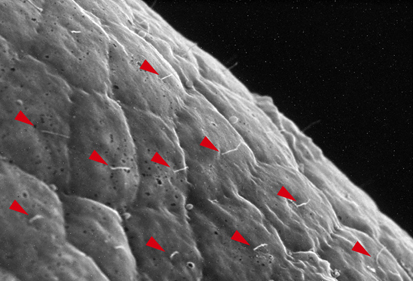
In many vertebrate embryos, monocilia-generated fluid flow in the node establishes left-sided mesodermal expression of nodal and breaks embryonic symmetry, which leads to the stereotypical left-right (LR) asymmetry seen in most animals. Tadpole larvae of the ascidian Halocynthia roretzi also show LR asymmetry but ascidian embryos have no cavity in which fluid flow can be generated. Now, Kazuhiko Nishide and colleagues show that in H. roretzi left-sided expression of nodal, triggered by neurula rotation, in the epidermis at the late neurula stage is required for LR asymmetry (p. 1467). Shortly before the onset of nodal expression, they report, the neurula rotates within the vitelline membrane until its left side is orientated downwards. Monocilia in the epidermis probably generate the driving force for this rotation. Moreover, experimental perturbation of neurula rotation indicates that contact between the left epidermis and the vitelline membrane generated through neurula rotation promotes left-sided nodal expression. Neurula rotation could also be involved in the establishment of LR asymmetry in other ascidian species, suggest the researchers.
Plus…
Diverse roles for VEGF-A in the nervous system
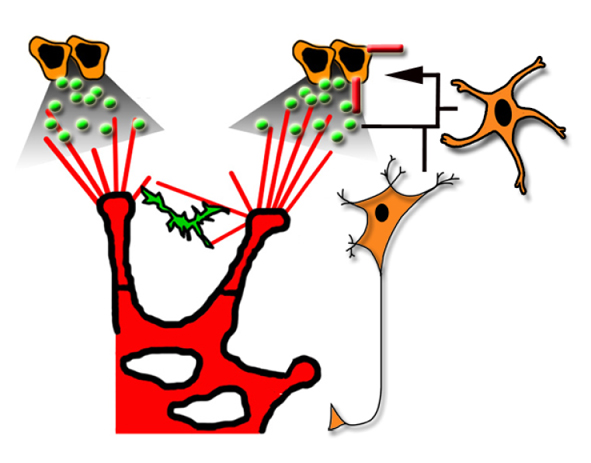
Recent studies, reviewed by Francesca Mackenzie and Christiana Ruhrberg, have shown that vascular endothelial growth factor A, which is best known for its roles in blood vessel growth, promotes a wide range of neuronal functions.
See the Review article on p. 1371
 (1 votes)
(1 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (8 votes)
(8 votes) Paul O’Neill shared some
Paul O’Neill shared some