Light sheet microscopy 101: Get started with a short video protocol
Posted by IchaJaroslav, on 13 April 2016
Here you can find out more about our video protocol on using light sheet microscopy to image zebrafish eye development.
Light sheet fluorescence microscopy has quickly become a popular technique in developmental biology. This method is very gentle to the samples, with fast acquisition speed and allows capturing the samples from any angle or from multiple angles at the same time (so called multi view imaging) (Stelzer, 2015). Such multi view imaging overcomes the degradation of the signal in Z axis and allows imaging of large specimens with high and almost isotropic resolution. The fact that light sheet microscopy is trending was confirmed when it was voted the method of the year 2014 by Nature Methods.
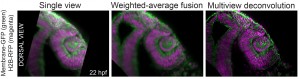
The major obstacle to mainstream adoption of this method was until recently the technical complexity for many biologists without a background in optics and computer science (Reynaud et al., 2015). The good news is that things have started to change and performing successful light sheet experiments is getting easier. For some time now, there are commercially available light sheet microscopes, which are easy to operate and the software solutions are not lagging behind, with plugins to process the resulting datasets as clickable GUIs (Amat et al., 2015; Preibisch et al., 2014; Preibisch et al., 2010) or Zeiss own solution in ZEN software. One word of warning, your hardware still has to be prepared to handle large volumes of data.
We recently contributed a video protocol (Icha, Schmied et al., 2016) to document a versatile light sheet microscopy experimental pipeline using a commercial microscope and an open source software solution for data processing. Specifically, we used the Lightsheet Z.1 microscope from Zeiss and the Multiview reconstruction application in Fiji to process the data. We demonstrated our approach by imaging several stages of retinal development in zebrafish. The general application for our pipeline would be long-term time-lapse imaging of morphogenetic processes during development using single or multi view acquisition. The protocol will take you through the essential steps of a light sheet microscopy experiment from mounting the sample to processing the data. The most complicated step is not taking the images, but the subsequent combination of image information from multiple views together. The solution we use is embedding fluorescent beads around the sample to register the different views onto each other and thereby to reconstruct the imaged volume of the sample.
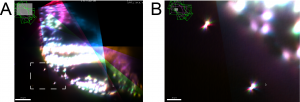
In the associated text, you will find a step-by-step protocol including all the experimental details plus troubleshooting in the discussion section. This should be especially useful for the newcomers to the light sheet fluorescence microscopy field.
This video protocol was created as a collaboration between the Norden lab (twitter @NordenLab) and the Tomancak lab (twitter @PavelTomancak) at MPI-CBG in Dresden. Enjoy and don’t hesitate to contact us in case you have questions. The full video is also available on the Norden lab website.
Other useful links:
EMBO course in Light sheet microscopy 2016
SPIM – Light Sheet Microscopy Literature Database
Open source hardware: DIY light sheet microscopes
Open SPIM wiki page
Open SPIN microscopy
Nature Methods method of the year 2014 thematic issue
References:
Amat, F., Höckendorf, B., Wan, Y., Lemon, W. C., McDole, K. and Keller, P. J. (2015). Efficient processing and analysis of large-scale light-sheet microscopy data. Nat Protoc 10, 1679–1696.
Icha, J., Schmied, C., Sidhaye, J., Tomancak, P., Preibisch, S., Norden, C. (2016). Using Light Sheet Fluorescence Microscopy to Image Zebrafish Eye Development. J Vis Exp 110, e53966.
Preibisch, S., Amat, F., Stamataki, E., Sarov, M., Singer, R. H., Myers, E. and Tomancak, P. (2014). Efficient Bayesian-based multiview deconvolution. Nat Meth 11, 645–648.
Preibisch, S., Saalfeld, S., Schindelin, J. and Tomancak, P. (2010). Software for bead-based registration of selective plane illumination microscopy data. Nat Meth 7, 418–419.
Reynaud, E. G., Peychl, J., Huisken, J. and Tomancak, P. (2015). Guide to light-sheet microscopy for adventurous biologists. Nat Meth 12, 30–34.
Stelzer, E. H. K. (2015). Light-sheet fluorescence microscopy for quantitative biology. Nat Meth 12, 23–26.


 (4 votes)
(4 votes)