Establishing connections
Posted by Bert De Rybel, on 16 December 2020
By Jos Wendrich, Yvan Saeys and Bert De Rybel
Nutrients such as phosphate are limiting in most soils. This has a great impact on crop yield in agriculture. Approaches to overcome these challenges often include using fertilizers, that in turn have a detrimental effect on the environment and our climate. Plants have developed adaptation mechanisms to efficiently forage the soil for phosphate such as modifying root system architecture and root hair density. In Wendrich et al., we unexpectedly identified a vascular transcription factor complex that controls root hair formation in response to limiting phosphate conditions.
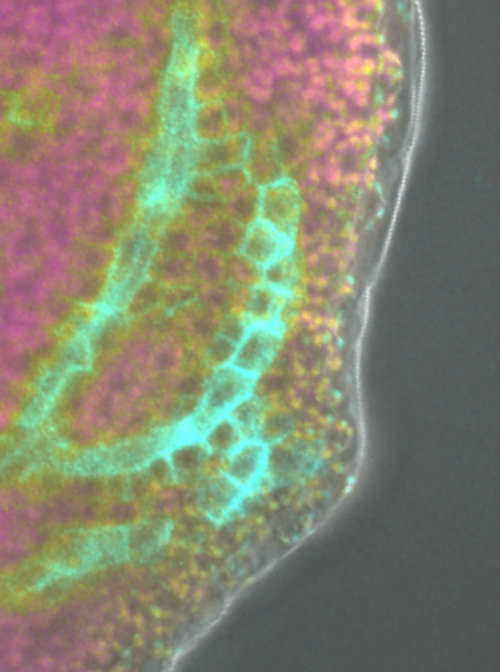
One of the main interests of our lab is identifying the molecular pathways controlled by the TARGET OF MONOPTEROS 5 and LONESOME HIGHWAY (TMO5/LHW) transcription factor heterodimer. In the root meristem, this complex is active in the inner most cells of the vasculature called the xylem. Here it controls the expression of LONELY GUY 4, which encodes an enzyme that is rate-limiting for the conversion of the phytohormone cytokinin into its bioactive form. Cytokinin is involved in many aspects of vascular development including patterning and proliferation. Using bulk transcriptomics, we previously revealed 273 target genes of the TMO5/LHW complex, but given the downstream intermediate cytokinin is mobile, we expected that many of these target genes would be expressed outside of the xylem cells. This is where this project started.
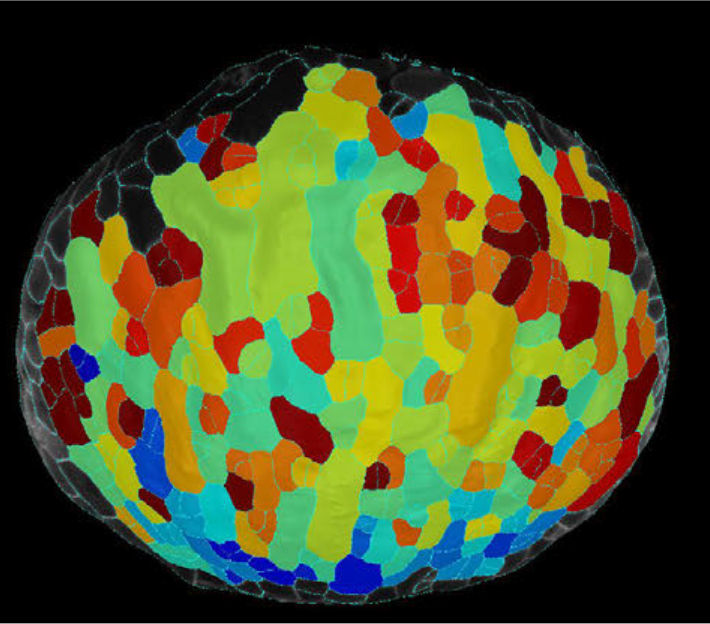
At that time, single-cell technologies were quickly becoming important in other fields and proved ideal to determine gene expression at unprecedented resolution. We thus decided we needed to get this technology up and running to answer our research question. Our institute had recently invested heavily to establish the technique, with commercial microfluidics systems and custom bioinformatic pipelines, but it remained unclear how reliable and efficient this would work on isolated plant cells. We teamed up with the group of Yvan Saeys in our neighboring VIB research center to generate a reference dataset for the Arabidopsis root meristem. We first needed to optimize the workflow leading up to the microfluidics single cell droplet emulsion. With their tough cell walls, plant cells are not easily dissociated from their tissue, requiring enzymatic digestion of the cell walls, called “protoplasting”. Cells in the outer layers are more accessible for these enzymes and tend to be overrepresented after in protoplast suspensions. We therefore first made sure that our single cell protoplast suspensions contained a good representation of each cell type, inner and outer layers, and was free of most debris and impurities, such as dead cells.
Unfortunately, this was the easy part. Now came the analysis, annotation and, importantly, full validation of the technology. This was where our collaboration effort truly started to pay off. The analysis pipelines were custom redesigned for our plant samples. This included building a visualization tool, for quick access to the data. Endless discussions, quick face-to-face check-ups and back-and-forth emailing ensured that we quickly learned from each other and from the dataset. Because of the rich resources available for Arabidopsis, we were able to annotate the main cell types of the root such as epidermis, ground tissues and vascular tissues. We however decided to take it a step further and zoom in to the level of sub-cell-types. Especially the different cell types of the vascular system were more difficult to annotate, as several cell types, like for example the procambium, only have limited known marker genes that are specifically expressed. Sub-cell-type predictions were confirmed by cell ploidy analysis and an extensive trajectory inference. By iteratively applying these trajectory methods, and having an open communication line between wet- and drylab, we were able to assess the robustness of the predicted trajectories and the resulting predicted subcluster annotations.
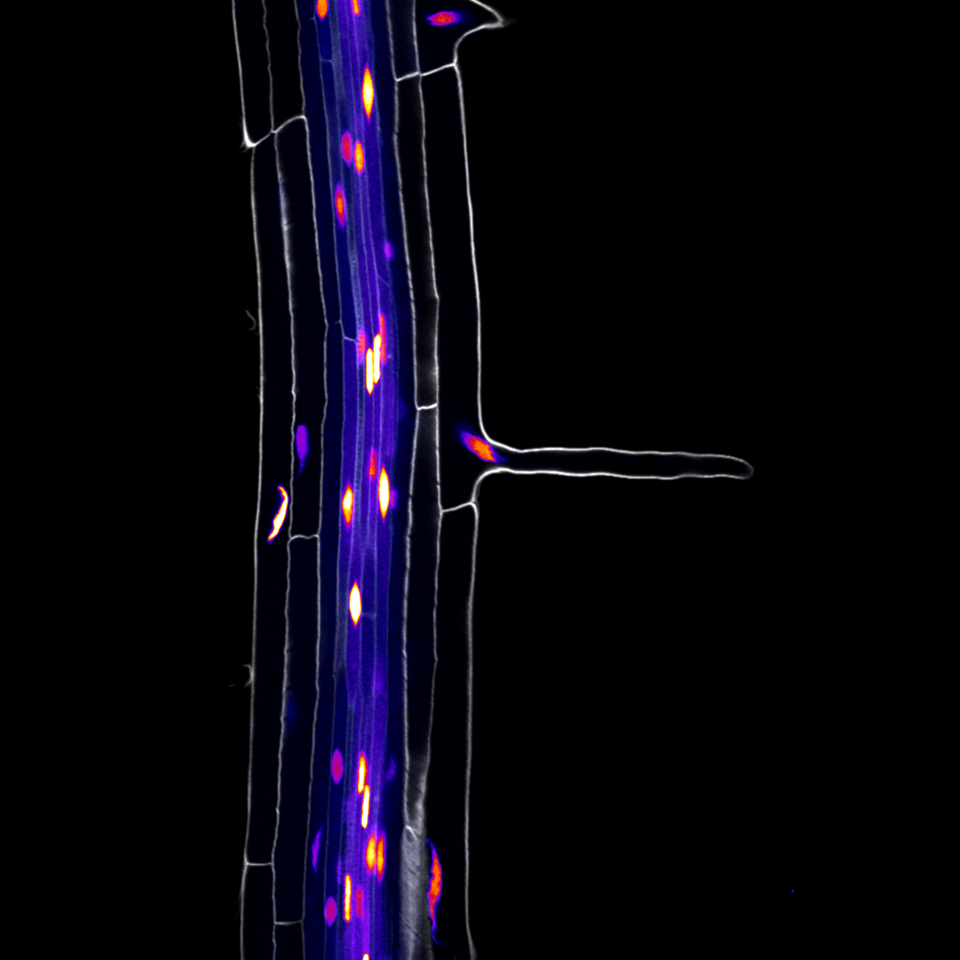
We next set out to evaluate the predictive power of this dataset, by construction of promoter reporter lines of genes with hitherto unknown expression patterns. All group members chipped in and a remarkable 100% of the lines with stable expression patterns validated the predicted cell type and developmental states. We were thus eager to apply this rich dataset on the 273 target genes of the TMO5/LHW complex previously identified. We identified an unexpected overrepresentation of genes expressed in root hairs (trichoblasts), suggesting that these vascular transcription factors may play a role in the development of the outermost trichoblast cells. Indeed, when overexpressing TMO5 and LHW in all cells of the root meristem, this dramatically increased the density of root hairs, resembling wild type roots grown in phosphate limiting conditions. Extensive follow-up genetic work allowed us to conclude that cytokinin, produced in inner vascular cells, acts as a mobile intermediate to control root hair response to low phosphate conditions in the outer most epidermal cell.
In our opinion, this is a good example of how collaborations within and between teams with different expertise can lead to new biological insights.


 (1 votes)
(1 votes)
 (No Ratings Yet)
(No Ratings Yet)
 (7 votes)
(7 votes)