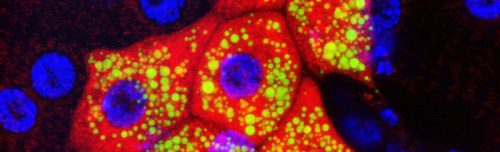
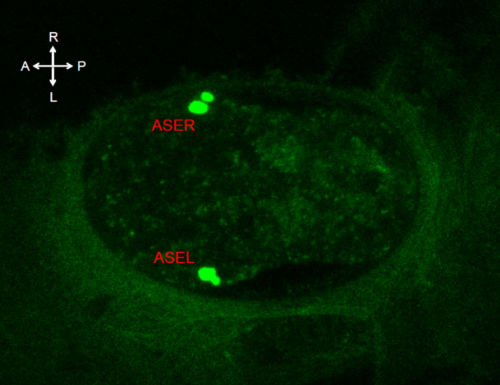
Development at the single cell level
Posted by the Node, on 17 July 2019
Development recently published a bumper Special Issue devoted to single cell approaches to developmental biology. A multitude of model systems featured – from Dicty to Drosophila to mouse to zebrafish – and the issue’s Reviews, Spotlight and Hypothesis gave an overview of the field’s current challenges and opportunities.
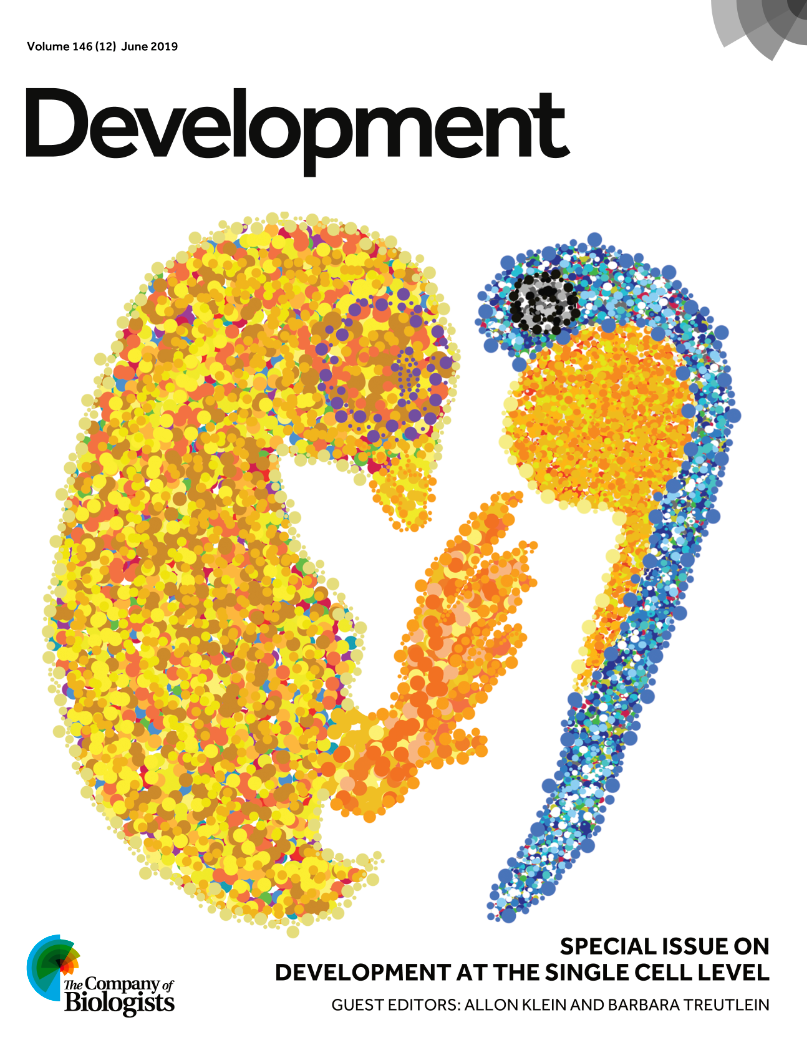
The cover was chosen by guest editors
Recently, two iconic developmental biology models entered into the single cell genomics era: chick and zebrafish. In this image, line art was traced using real embryo images for reference and filled with individual dots to represent the reduction of the whole embryo to its smallest structural, functional and biological unit: the cell.
The Guest Editors also surveyed the field and the issue in their Editorial, which we reprint here:
Single cell analyses of development in the modern era
Single cell analyses encompass multiple approaches that, we suggest, can be summarized by two unifying goals: to explain the cellular composition of tissues; and to characterize the dynamic processes of cells. Both forms of investigation have an old history, but have undergone rapid transformation in the past decade. This Special Issue of Development offers us an opportunity to reflect on the state of this rapidly evolving field.
Arguably, single cell analysis as we recognize it today became established after the broad acceptance of cell theory in the late 19th century. Cells were first recognized much earlier by Robert Hooke and Anton van Leeuwenhoek in the 17th century, enabled by technological development (the microscope) and by their curiosity about how a macroscopic biological system could be understood in terms of its constituent parts. Since the early studies of Hooke and van Leeuwenhoek, the tools of single cell analysis have been continuously evolving, driven by innovations in technology, sample preparation and new questions. By the end of the 19th century, cell theory was firmly established following significant improvements in microscopy and in biochemical techniques that could distinguish between morphologically similar cells and separate them into distinct states by fractionation or staining. By the late 20th century, the development of monoclonal antibodies had rapidly expanded the repertoire of tools by which biologists could distinguish cellular states. Live imaging methods, and the development of fluorescent proteins, extended the study of dynamic molecular processes in cells. By the start of the 21st century, differences between cells could be measured with single molecule accuracy, allowing the study of stochastic chemistry within single cells, and accelerating a new field that aimed to understand information processing and noise in biological systems. In parallel, molecular methods developed in the 1990s allowed the labeling of subsets of cells using transgenic approaches, establishing the fate mapping of cells as an important gold standard technique for determining developmental relationships. Thus, single cell techniques have driven biological discovery for centuries.
Over the past two decades, however, technological advances have revolutionized the scale and complexity of single cell analyses. Starting in a few labs and then spreading rapidly, single cell investigation merged with genome-scale analytical methods. The most mature of these technologies today is single cell RNA-sequencing, which can measure the expression profile of thousands of genes simultaneously in thousands of cells (and more) in a single experiment. In parallel, innovations in microscopy now allow us to track single cells in complex tissues with minute-by-minute time resolution and with minimal phototoxicity, spherical aberration or signal attenuation. Some of these technologies are now widely accessible and are being implemented across developmental biology. The new resolution afforded by these methods has led to multiple discoveries in a short period of time, including the discovery of novel cell types, revisions to established differentiation hierarchies and identification of novel regulators of fate choice. In stem cell biology, these measurements allow the analysis of in vitro differentiation products, and the benchmarking of stem cell-derived cells against their natural counterparts. In evolutionary biology, they allow orthologous cell types to be compared across species. In regenerative biology, they allow the stem and progenitor cells driving tissue recovery, as well as the behavior of supporting immune and stromal cells, to be identified. Across different fields, single cell genomic atlases of tissues offer new perspectives on cell types and cell states. In parallel, single cell live-imaging approaches have challenged established mechanistic perspectives of molecular signal transduction, chromatin modification and transcription factor dynamics.
While modern single cell analyses are accelerating biological discovery, we note that they have also imposed a new demand on researchers seeking to benefit from these tools. Unlike single cell innovations of the previous century, modern single cell analytical methods produce high-dimensional data, or ‘big data’, in the sense that raw data obtained using these methods cannot be inspected without the aid of computational algorithms for dimensionality reduction. Although data of a statistical nature is not new to developmental biology, the examination of cell states over tens of thousands of genes cannot be analyzed using basic visualization methods such as histograms. Even seemingly ‘simple’ representations using heatmaps often undergo multiple steps of normalization and hierarchical clustering prior to visualization. Similar challenges occur in visualizing cell behaviors across space and time, in relation to complex spatial neighborhoods or patterns of protein dynamics. Fortunately, there are now many algorithms available to visualize and interpret single cell data. Yet each makes particular assumptions that may skew interpretation of the data, and each is dependent on parameters that can alter the results. Furthermore, many algorithms are non-deterministic, leading to variable representations of the same data. We propose that a unique aspect of modern single cell analysis is its demand for computational methods. These methods offer opportunities for collaboration and immersion in computational biology, and may be altering the training of developmental biologists.
With this background, we are excited to present several review-based articles in this Special Issue that survey the ways in which modern single cell analytical techniques and computational methods are driving advances in developmental biology. We are also delighted by a strong representation of primary research papers, which provide examples of many of the reviewed concepts and how they drive discovery.
Our Reviews broadly cover the conceptual challenges resulting from single cell analysis, the new methods that are available and some of the successes of single cell analyses to date. Two separate articles challenge us to consider how we should understand the notion of cell identity and cell type. Bo Xia and Itai Yanai (Xia and Yanai, 2019) propose that differences between cell types in an organism follow an understandable pattern that allows them to be organized into a ‘periodic table’ that reveals the logic of their underlying structure. Samantha Morris (Morris, 2019) proposes a complementary view that defines cells in terms of their lineage, molecular state and functional phenotypes. In doing so, she importantly clears up some of the nomenclature in the field. Both perspectives highlight the new opportunities that have emerged from the unifying, quantitative language of single cell analyses that force us to confront definitions that have emerged in various isolated studies of distinct tissues. Two additional Reviews survey the use of single cell analyses to reconstruct developmental dynamics. Fabian Theis and colleagues (Tritschler et al., 2019) review the methods available to infer dynamic trajectories from high-dimensional single cell data, while Aaron McKenna and James Gagnon (McKenna and Gagnon, 2019) survey cutting-edge technologies that combine single cell analyses with lineage tracing, thus building bona fide dynamic information into static snapshots. Then three further Reviews provide insights into the future beyond the mature techniques of single cell transcriptomics. Connor Ludwig and Lacramioara Bintu (Ludwig and Bintu, 2019) survey the multiple modalities of measurements beyond RNA-seq, focusing on single cell analyses of chromatin state. Prisca Liberali and colleagues (Mayr et al., 2019) discuss spatially resolved single cell analyses. Finally, Pulin Li and Michael Elowitz (Li and Elowitz, 2019) discuss the use of single cell live imaging to learn about information processing by cells.
Putting these ideas into play, the research papers in this Special Issue demonstrate a range of single cell techniques and the discoveries that they enable. Two papers apply live imaging to demonstrate the role of a signal transduction pathway in differentiation (Deathridge et al., 2019) or to demonstrate stochasticity in fate choice (Antolović et al., 2019); several papers build transcriptomic maps of specific developing tissues that will prove useful resources and reveal new hypotheses for developmental regulation (Combes et al., 2019; Martin et al., 2019; Hulin et al., 2019; Li et al., 2019a,b); several studies computationally reconstruct developmental dynamics (Bastidas-Ponce et al., 2019; Delile et al., 2019; Guo and Li, 2019; Prior et al., 2019; van Gurp et al., 2019); and one study benchmarks in vitro differentiation to in vivo development and develops new computational tools to do so (Edri et al., 2019). Taking a slightly different approach, other papers use single cell analyses to characterize cell behaviors during morphogenesis (Amini et al., 2019; Yang et al., 2019), to investigate how cells respond to DNA or tissue damage (Miermont et al., 2019; Dell’Orso et al., 2019), and to understand how heterogeneities in gene and protein concentration influence development (Papadopoulos et al., 2019; Reznik et al., 2019; Velte et al., 2019).
Approaches to single cell analysis began three and a half centuries ago with improvements in microscopy. Today’s modern approaches, although hard to relate to those of 17th century Europe, echo this first revolution. Then, as now, continuous innovations in technology have led to sudden and widespread acceleration in biological inquiry. As this Special Issue illustrates, methods that once required specialized expertise are becoming widely accessible. Yet rapid innovation still continues. Will the tools and concepts surveyed in this Special Issue still be relevant 5 years from today? As you consider this question, we would very much like to thank the authors and referees of the articles in this Special Issue for their contributions, and we hope you enjoy reading it!


 (1 votes)
(1 votes) (No Ratings Yet)
(No Ratings Yet)