Decoding light for developmental timing
Posted by shruti.vemaraju, on 13 June 2019
Shruti Vemaraju¹ and Richard A. Lang¹-²
¹Center for Chronobiology,¹The Visual Systems Group, Abrahamson Pediatric Eye Institute, Division of Pediatric Ophthalmology, ²Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229, USA. ²Department of Ophthalmology, University of Cincinnati, College of Medicine, Cincinnati, OH 45229, USA
We live on a planet that is close to a yellow dwarf star. This means that throughout the evolution of life, photons have been available. Living systems have exploited photon availability for adaptive advantage using pathways like photosynthesis, the circadian clock and the visual system. However, in addition to these obvious examples, we are learning that metazoans can decode light information in other ways. Here we comment on recent work from the Lang lab that describes how light signals can provide developmental timing cues.
The embryonic eye has an elaborate vessel network, called the hyaloid vasculature, that supplies nutrients and oxygen to the developing retina. This network is transient and undergoes programmed regression concomitantly with the development of retinal vasculature. Regression of the hyaloid vessels is an adaptation for high acuity vision: Since the structure lies in the optical path and would scatter light, its regression is necessary for optical function of the eye. The clinical conditions persistent fetal vasculature (PFV) and persistent hyperplastic primary vitreous (PHPV) are a consequence of abnormal hyaloid vessel regression.
Research in the Lang lab has focused on development of vasculature in the eye since 1993 when we showed that myeloid cells play an important role in scheduled vascular regression (Lang and Bishop, 1993). The molecular pathways required for hyaloid vessel regression (the Wnt, Angiopoietin and VEGF pathways) have been defined by our group and by several others (Gale et al., 2002; Glass II et al., 2005; Kurihara et al.; Liu and Nathans, 2008; Lobov et al., 2002; Nayak et al., 2018; Rao et al., 2007; Yoshikawa et al., 2016). The hyaloid vessel system is an interesting subject of study in part because it can be dissected from the eye as a whole mount and the vessels easily counted. The hyaloid vasculature may seem like an obscure structure (it is) but has featured on the front cover of Development twice (Nayak et al., 2018; Rao et al., 2007)(Fig. 1).

Despite all that had been learned, the trigger that initiated hyaloid regression remained to be elucidated. When the Lang lab considered this question, we thought about the possibility that light stimulation could be the trigger. After all, this was an eye, a structure designed to respond to light, and hyaloid vessel regression began soon after birth when light exposure levels would presumably be elevated. To investigate this initially, we (that is, Lang Lab alumnus Sujata Rao) dark-reared mice and found hyaloid vessel persistence, an indication that light stimulation could be involved.
What unfurled next was a nice illustration of how science investigations sometimes work. Our colleague Maureen McCall was visiting from Louisville and, after she heard about our work, recommended we contact David Copenhagen (UCSF) because he was also working on light response pathways in neonatal mice. Excited by the possibilities, we contacted David and learned that newborn mice have a negative phototaxis (a light aversion response) mediated by melanopsin (OPN4), one of the non-canonical retinal opsins (Johnson et al., 2010). This raised the possibility that melanopsin might also regulate hyaloid regression and sure enough, when Sujata assessed Opn4 null mice, we found hyaloid vessel persistence (thanks for the hot tip Maureen). Subsequently, the Lang and Copenhagen labs worked together to understand how melanopsin controlled the timing of vascular regression.
Key findings included the observation that dark reared and Opn4 null mice developed an over-abundance of retinal neurons, showed elevated oxygen demand and elevated levels of vascular endothelial growth factor A (VEGFA), a crucial promoter of angiogenesis and vascular endothelial cell survival (Rao et al., 2013). Elevated levels of VEGFA provided a good explanation for the promiscuous retinal angiogenesis and hyaloid vessel persistence characteristic of the Opn4 null and dark-reared mice (Rao et al., 2013). In a little bit of a surprise, the analysis suggested that the light-OPN4 pathway functioned before birth in a fetal eye that was directly light responsive. While some of our colleagues have struggled with the idea that deep tissue photoreceptors can receive sufficient light to function, there is evidence throughout the animal kingdom that this is one mode of light detection (Miyashita et al., 1996; Nakane et al., 2010; Okano et al., 1994) and recent analysis from the Lang and Copenhagen labs confirms that this is the way this pathway works (stay tuned….).
Prompted by this work, the Lang lab become much more curious about the possibility that other light response pathways could play an important role in development. Genome sequencing nearly two decades earlier had identified two other non-canonical opsins, neuropsin (OPN5, (Tarttelin et al., 2003)) and encephalopsin (OPN3, (Blackshaw and Snyder, 1999)). Both of these were expressed in the retina and so as a first assessment, the Lang lab determined whether a null mouse for either opsin has a vascular phenotype in the eye: In Opn5 null mice, we found one (Nguyen et al., 2019).
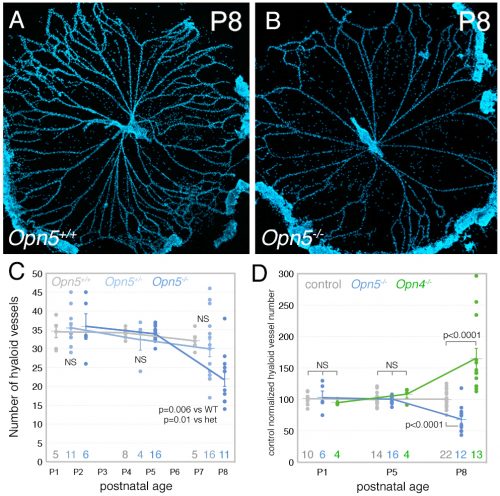
The Lang lab has trained a small cadre of researchers to perform hyaloid vessel dissections (the dissection requires steady hands, patience, specialized technique, and freshly harvested eyes). On the day we first analyzed the Opn5 mouse hyaloid vessels, we had three litters all at the same stage of development. This meant a long day of dissections for Postdoc Shruti Vemaraju (50 eyes and about 6 hours of dissection) in which she was blinded to the genotype and so had no idea of the outcome as she worked. After all the staining and counting was complete, we were excited to learn that loss of OPN5 function resulted in precocious hyaloid vessel regression (Nguyen et al., 2019). This was unusual because all hyaloid phenotypes described to date showed vessel persistence (Fig. 2). OPN5 is known to be stimulated by violet light with peak absorbance at 380 nm (Kojima et al., 2011). Withdrawing this wavelength during the neonatal period mimicked the precocious hyaloid vessel regression seen with loss of OPN5 function (Nguyen et al., 2019). This made a strong argument that OPN5 was functioning as an opsin to regulate hyaloid regression.
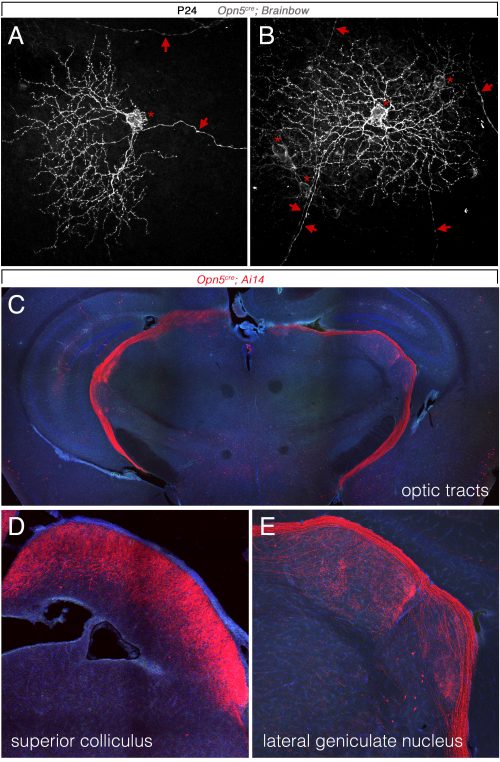
So, to summarize, we now knew that two distinct opsins, OPN4 and OPN5 each regulated hyaloid vessel regression, but because the phenotypes were opposite, probably used distinct mechanisms. We knew from prior work (a collaboration with the labs of Russell Van Gelder, King-Wai Yau and Ethan Buhr) that Opn5 was expressed in retinal ganglion cells (RGCs)(Buhr et al., 2015). Since we had not been able to identify any antibodies that detected OPN5, we generated an Opn5cre allele as a means of targeting and characterizing Opn5 RGCs. Conversion of the Ai14 tdTomato cre reporter confirmed that positive cells had all the characteristics of RGCs (Fig. 3) including projections to the lateral geniculate nucleus and superior colliculus.
When Lang lab Research Associate Minh-Thanh Nguyen was characterizing the Opn5 null retina, she noticed an unusual pattern of expression of tyrosine hydroxylase, the rate limiting enzyme in the pathway that synthesizes dopamine. This suggested that dopamine might be changed and, according to an ELISA assessment, it was low in retinal neurons and high in the extracellular vitreous (where the hyaloid vessels reside). This was interesting because dopamine can have an anti-vascular function: via the dopamine receptor DRD2, dopamine can activate the phosphatase Shp2 and promote dephosphorylation of VEGF receptor 2 (Sinha et al., 2009). This suggested the hypothesis that an OPN5 light response pathway regulated hyaloid vessel regression using dopamine as a signaling intermediate.
Aided by dopamine expert and collaborator Mike Iuvone at Emory University, we investigated this idea further and showed that hyaloid regression could be both positively (precocious regression with receptor agonists) and negatively (vessel persistence with receptor antagonists) regulated by dopamine signaling. We further showed that a key regulator of dopamine uptake, the dopamine transporter (DAT) was activated (via phosphorylation (Foster et al., 2012) by 380 nm light in an Opn5-dependent manner. Phospho-DAT was detected in a network of neuronal processes throughout the inner retina and so was well placed to globally regulate dopamine levels. Using a pharmacological inhibitor of DAT, we could revert hyaloid vessel persistence resulting from dark-rearing to normal numbers and also reproduce an Opn5 null precocious hyaloid vessel phenotype in Opn5 heterozygote mice. These findings led to the hypothesis that hyaloid vessel regression is regulated by an intricate balance between dopamine and VEGFA signaling.
The hypothesis that dopamine was a signaling intermediate also required that hyaloid vascular endothelial cells (VECs) expressed a dopamine receptor. We detected DRD2 in the hyaloid vessels by immunolabeling and further showed that conditional deletion of a Drd2 floxed allele in VECs resulted in hyaloid persistence. When combined with Opn5 loss-of-function (elevated dopamine levels in vitreous, precocious regression), deletion of Drd2 from VECs abrogates the effects of dopamine signaling and restores hyaloid vessel numbers to normal. This was consistent with a direct effect of dopamine promoting hyaloid vessel regression.
A further prediction of the hypothesis was that VEGFR2 signaling in the hyaloid vessels would be influenced by dopamine. To test this, Lang lab Postdoc Yoshinobu Odaka performed immunoblotting on the vanishingly small quantities of cell lysates available from hyaloid vessel preparations. He assessed the levels of total and phospho–tyrosine 1173-VEGFR2 (an activating modification) in control and Drd2 VEC conditional deletion hyaloids. Consistent with the hypothesis, pY1173-VEGFR2 was elevated when Drd2 was deleted. This supported the idea that normally DA signaling suppresses VEGFR2 and provided a mechanistic explanation for how the high levels of vitreal dopamine in the Opn5 null could promote precocious hyaloid vessel regression.
During review of the manuscript describing this work, we were asked quite a few questions about how the OPN4 and OPN5 vascular pathways were integrated. The schematic (Fig. 4) describes our current model. We propose that blue light photons activate OPN4 before birth. Through a mechanism not currently understood, this suppresses the number of retinal neurons that develop. When mice are dark–reared or have OPN4 loss-of-function, cellularity and oxygen demand are higher, and this stimulates production of VEGFA. Under those abnormal conditions, VEGFA within the retina produces promiscuous angiogenesis while vitreal VEGFA suppresses regression of the hyaloid vessels. This means that normally, blue light stimulation of fetal retina suppresses the levels of VEGFA as a prerequisite for regression of the hyaloid vessels.
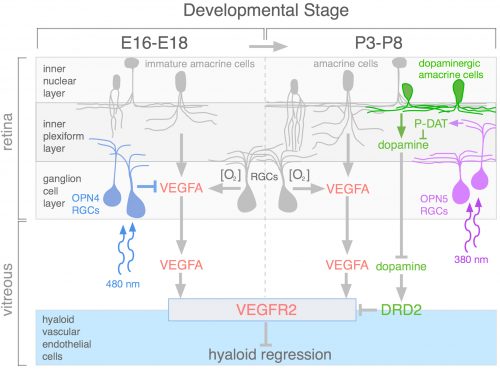
Current analysis suggests that OPN5 functions after birth to suppress the level of vitreal dopamine. This dopamine suppresses VEGFR2 signaling and promotes hyaloid vessel regression. However, this occurs when dopamine levels in the eye are generally rising because the TH+ amacrine cells that are the source of dopamine are differentiating and becoming active. This series of positive and negative influences means that the net effect of violet light and OPN5 activity is to sustain the hyaloid vessels. This mechanism likely evolved to ensure that hyaloid vessel regression does not occur before the first layer of retinal vasculature is complete: this would prevent hypoxia by ensuring that one of the vascular networks was always functional. One way to think about OPN4 and OPN5 function in the developing eye is that they are the detectors for developmental timing cues. A reminder that in development, timing is everything.
References
Blackshaw, S. and Snyder, S. H. (1999). Encephalopsin: a novel mammalian extraretinal opsin discretely localized in the brain. J Neurosci 19, 3681–3690.
Buhr, E. D., Yue, W. W., Ren, X., Jiang, Z., Liao, H. W., Mei, X., Vemaraju, S., Nguyen, M. T., Reed, R. R., Lang, R. A., et al. (2015). Neuropsin (OPN5)-mediated photoentrainment of local circadian oscillators in mammalian retina and cornea. Proc Natl Acad Sci U S A 112, 13093–13098.
Foster, J. D., Yang, J. W., Moritz, A. E., ChallaSivaKanaka, S., Smith, M. A., Holy, M., Wilebski, K., Sitte, H. H. and Vaughan, R. A. (2012). Dopamine transporter phosphorylation site threonine 53 regulates substrate reuptake and amphetamine-stimulated efflux. J. Biol. Chem. 287, 29702–29712.
Gale, N. W., Thurston, G., Hackett, S. F., Renard, R., Wang, Q., McClain, J., Martin, C., Witte, L., Witte, M. H., Jackson, D., et al. (2002). Angiopoietin-2 is Required for Postnatal Angiogenesis and Lymphatic Patterning, and Only the Latter is Rescued by Angiopoietin-1. Dev Cell 3, 411–423.
Glass II, D. A., Bialek, P., Ahn, J. D., Starbuck, M., Patel, M. S., Clevers, H., Taketo, M. M., Long, F., McMahon, A. P., Lang, R. A., et al. (2005). Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev. Cell 8,.
Johnson, J., Wu, V., Donovan, M., Majumdar, S., Renteria, R. C., Porco, T., Van Gelder, R. N. and Copenhagen, D. R. (2010). Melanopsin-dependent light avoidance in neonatal mice. Proc Natl Acad Sci U S A 107, 17374–17378.
Kojima, D., Mori, S., Torii, M., Wada, A., Morishita, R. and Fukada, Y. (2011). UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS One 6, e26388.
Kurihara, T., Kubota, Y., Ozawa, Y., Takubo, K., Noda, K., Simon, M. C., Johnson, R. S., Suematsu, M., Tsubota, K., Ishida, S., et al. von Hippel-Lindau protein regulates transition from the fetal to the adult circulatory system in retina. Development 137, 1563–1571.
Lang, R. A. and Bishop, J. M. (1993). Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell 74,.
Liu, C. and Nathans, J. (2008). An essential role for frizzled 5 in mammalian ocular development. Development 135, 3567–76.
Lobov, I. B., Brooks, P. C. and Lang, R. A. (2002). Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. PNAS 99, 11205–11210.
Miyashita, Y., Moriya, T., Yokosawa, N., Hatta, S., Arai, J., Kusunoki, S., Toratani, S., Yokosawa, H., Fujii, N. and Asami, K. (1996). Light-sensitive response in melanophores of Xenopus laevis: II.Rho is involved in light-induced melanin aggregation. J. Exp. Zool. 276, 125–31.
Nakane, Y., Ikegami, K., Ono, H., Yamamoto, N., Yoshida, S., Hirunagi, K., Ebihara, S., Kubo, Y. and Yoshimura, T. (2010). A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc. Natl. Acad. Sci. 107, 15264–15268.
Nayak, G., Odaka, Y., Prasad, V., Solano, A. F., Yeo, E.-J., Vemaraju, S., Molkentin, J. D., Trumpp, A., Williams, B., Rao, S., et al. (2018). Developmental vascular regression is regulated by a Wnt/β-catenin, MYC and CDKN1A pathway that controls cell proliferation and cell death. Development 145, dev154898.
Nguyen, M.-T., Vemaraju, S., Nayak, G., Odaka, Y., Buhr, E. D., Alonzo, N., Tran, U., Batie, M., Upton, B. A., Darvas, M., et al. (2019). An Opsin 5-dopamine pathway mediates light-dependent vascular development in the eye. Nat. Cell Biol. In press,.
Okano, T., Yoshizawa, T. and Fukada, Y. (1994). Pinopsin is a chicken pineal photoreceptive molecule. Nature 372, 94–7.
Rao, S., Lobov, I. B., Vallance, J. E., Tsujikawa, K., Shiojima, I., Akunuru, S., Walsh, K., Benjamin, L. E. and Lang, R. A. (2007). Obligatory participation of macrophages in an angiopoietin 2-mediated cell death switch. Development 134,.
Rao, S., Chun, C., Fan, J., Kofron, J. M., Yang, M. B., Hegde, R. S., Ferrara, N., Copenhagen, D. R. and Lang, R. A. (2013). A direct and melanopsin-dependent fetal light response regulates mouse eye development. Nature 494, 243–246.
Sinha, S., Vohra, P. K., Bhattacharya, R., Dutta, S., Sinha, S. and Mukhopadhyay, D. (2009). Dopamine regulates phosphorylation of VEGF receptor 2 by engaging Src-homology-2-domain-containing protein tyrosine phosphatase 2. J Cell Sci 122, 3385–3392.
Tarttelin, E. E., Bellingham, J., Hankins, M. W., Foster, R. G. and Lucas, R. J. (2003). Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue. FEBS Lett 554, 410–416.
Yoshikawa, Y., Yamada, T., Tai-Nagara, I., Okabe, K., Kitagawa, Y., Ema, M. and Kubota, Y. (2016). Developmental regression of hyaloid vasculature is triggered by neurons. J. Exp. Med. 213, 1175–1183.



 (3 votes)
(3 votes)

 (No Ratings Yet)
(No Ratings Yet)













