Here are the highlights from the new issue of Development:
Glucocorticoid and STAT3: tipping the balance in the lung

The epithelial cells found at the distal tips of the developing lung comprise a multipotent progenitor population. During development, these cells first give rise to bronchiolar cells, which form the conducting airways, but then switch to producing alveolar cells, which form the sites of gas exchange. Here, on. p. 3686, Emma Rawlins and co-workers investigate the factors that control this transition in the mouse lung. They report that distal tip progenitors begin to express alveolar fate markers at around E16.5. Using a grafting assay, the researchers reveal that extrinsic, rather than intrinsic, factors determine the fate of tip progenitors. Importantly, they reveal that the glucocorticoid and STAT3 signalling pathways operate in parallel to promote alveolar fate; both pathways are sufficient but not necessary for specifying alveolar cells. Finally, the authors demonstrate that STAT3 signalling is also active at a similar stage of lung development in humans. Overall, these results highlight that the fate of lung epithelial cells is controlled by extrinsic signalling from surrounding tissues, a finding that has important implications for developing therapies that can restore alveolar capacity in human lungs.
Mapping out testis formation

The mammalian testis contains male germ cells as well as a number of somatic cell types, including supporting cells (such as Sertoli cells) and interstitial cells (such as Leydig cells). Although the origin and differentiation of germ cells has been well-characterized, the developmental course of somatic lineages in the testis is ill-defined. Now, Humphrey Yao and colleagues construct a comprehensive map of somatic cell lineage progression in the mouse testis (p. 3700). Their lineage-tracing studies reveal that both supporting and interstitial cells arise from a population of WT1-expressing progenitors. A sub-population of these, marked by SOX9 expression, then gives rise to Sertoli cells of the testis cords. The researchers demonstrate that the interstitial progenitors further diversify, based on differential Notch and Hedgehog pathway activation, giving rise to foetal steroid-producing Leydig cells and non-steroidogenic progenitors. Finally, the authors report that non-steroidogenic progenitors, which are maintained in an undifferentiated state throughout foetal development, eventually become adult Leydig cells. Together, these findings provide key insights into the lineage progression events that occur during testis development in mammals.
YY1 invokes a gut (metabolic) reaction

Incomplete intestinal development is a common gastrointestinal complication in neonates, yet the factors that control the late stages of intestinal development are unclear. Here, Michael Verzi and colleagues uncover a key role for the transcription factor YY1 in intestinal morphogenesis in mice (p.3711). They demonstrate that Yy1 expression in the developing endoderm is required for the correct formation of villi – the structures that extend into the intestinal lumen. In particular, the extension of villi, rather than the initiation of villogenesis, is compromised in Yy1 mutants. Transcriptomic analyses reveal that genes associated with mitochondrial function are perturbed in Yy1 mutants. In line with this, the authors report that Yy1 loss leads to defective mitochondrial morphology. The researchers further demonstrate that oxidative phosphorylation genes are upregulated at the time of villus growth, and that mitochondrial inhibitors can block villus formation in explant cultures, suggesting that aerobic respiration is required for the late stages of intestinal development. Finally, the authors show that patients presenting with necrotizing enterocolitis, which is thought to be caused by incomplete intestinal development, exhibit reduced expression of YY1 target genes and oxidative phosphorylation genes. In summary, these findings highlight a clear link between metabolism and organogenesis.
Identifying active enhancers: FAIR(E) play

Tissue-specific control of gene expression is crucial during development. In recent years, a number of genome-wide approaches have been used to identify potential regulatory elements that control gene expression, but determining which of these are functionally relevant has been a challenge. Here, Stephen Crews and colleagues describe an approach to identify active and biologically relevant enhancers (p. 3723). They focus on gene expression in Drosophila CNS midline neurons, which are well-characterized with regards to their gene regulatory mechanisms and hence serve as a useful model for studying transcriptional regulation. The researchers use formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq) analysis of purified midline cells and compare this with whole embryo FAIRE data. Using this approach, the authors identify known enhancers as well as novel enhancers that act specifically in midline cells. They also compare midline FAIRE-seq data with currently available midline expression and enhancer datasets, and reveal, for example, that many genomic fragments that have previously been shown to drive midline expression are unlikely to function in vivo. Overall, this approach emphasizes the importance of using highly purified cells in genome-wide analyses and highlights potential limitations to using standard reporter assays for identifying bona fide enhancers.
PLUS:
Regulation and plasticity of intestinal stem cells during homeostasis and regeneration
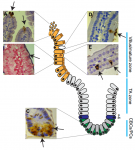 The intestinal epithelium is the fastest renewing tissue in mammals and has a large flexibility to adapt to different types of damage. Here, Joep Beumer and Hans Clevers review our current understanding of how intestinal stem and progenitor cells contribute to the homeostasis and regeneration of the intestine, highlighting the different signaling pathways that regulate their behavior. See the Review on p. 3639
The intestinal epithelium is the fastest renewing tissue in mammals and has a large flexibility to adapt to different types of damage. Here, Joep Beumer and Hans Clevers review our current understanding of how intestinal stem and progenitor cells contribute to the homeostasis and regeneration of the intestine, highlighting the different signaling pathways that regulate their behavior. See the Review on p. 3639
From the stem of the placental tree: trophoblast stem cells and their progeny
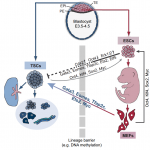 Trophoblast stem cells (TSCs) retain the capacity to self-renew indefinitely and harbour the potential to differentiate into all trophoblast subtypes of the placenta. Recent studies have shown how signalling cascades integrate with transcription factor circuits to govern the fine balance between TSC self-renewal and differentiation. In addition, breakthroughs in reprogramming strategies have enabled the generation of TSCs from fibroblasts. Here, Paulina Latos and Myriam Hemberger discuss these advances. See the Review on p. 3650
Trophoblast stem cells (TSCs) retain the capacity to self-renew indefinitely and harbour the potential to differentiate into all trophoblast subtypes of the placenta. Recent studies have shown how signalling cascades integrate with transcription factor circuits to govern the fine balance between TSC self-renewal and differentiation. In addition, breakthroughs in reprogramming strategies have enabled the generation of TSCs from fibroblasts. Here, Paulina Latos and Myriam Hemberger discuss these advances. See the Review on p. 3650
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 Finally, the Autum Meeting hosted the inaugural Dennis Summerbell Lecture which was given by
Finally, the Autum Meeting hosted the inaugural Dennis Summerbell Lecture which was given by  (1 votes)
(1 votes) Dementia causes enormous personal hardship and costs the UK ~£23 billion every year. The second most common form is Frontotemporal lobar degeneration (FTLD). About 40% of FTLD cases have genetic causes, with >8% involving abnormal aggregate-forming GA, GR, PR, GP and AP dipeptide repeat proteins (DPRs).
Dementia causes enormous personal hardship and costs the UK ~£23 billion every year. The second most common form is Frontotemporal lobar degeneration (FTLD). About 40% of FTLD cases have genetic causes, with >8% involving abnormal aggregate-forming GA, GR, PR, GP and AP dipeptide repeat proteins (DPRs).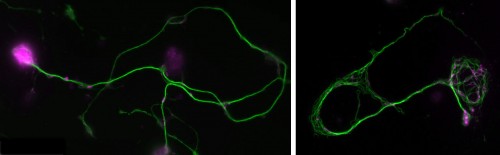
 ch of a new Special Collection named Spotlight on Rat: Translational Impact. The rat is a key model for basic and preclinical studies of physiology, pharmacology, toxicology and neuroscience, underlining its importance in studies of human disease. There are many reasons for its suitability as a model system – the close evolutionary and genomic relationship to humans, the sophistication and sociability of the animal, the ease of physiological and behavioural measurements, and the recent proliferation of transgenic and knockout rats, enabled by new and improved technologies for genetic manipulation.
ch of a new Special Collection named Spotlight on Rat: Translational Impact. The rat is a key model for basic and preclinical studies of physiology, pharmacology, toxicology and neuroscience, underlining its importance in studies of human disease. There are many reasons for its suitability as a model system – the close evolutionary and genomic relationship to humans, the sophistication and sociability of the animal, the ease of physiological and behavioural measurements, and the recent proliferation of transgenic and knockout rats, enabled by new and improved technologies for genetic manipulation.



 The intestinal epithelium is the fastest renewing tissue in mammals and has a large flexibility to adapt to different types of damage. Here,
The intestinal epithelium is the fastest renewing tissue in mammals and has a large flexibility to adapt to different types of damage. Here,  Trophoblast stem cells (TSCs) retain the capacity to self-renew indefinitely and harbour the potential to differentiate into all trophoblast subtypes of the placenta. Recent studies have shown how signalling cascades integrate with transcription factor circuits to govern the fine balance between TSC self-renewal and differentiation. In addition, breakthroughs in reprogramming strategies have enabled the generation of TSCs from fibroblasts. Here,
Trophoblast stem cells (TSCs) retain the capacity to self-renew indefinitely and harbour the potential to differentiate into all trophoblast subtypes of the placenta. Recent studies have shown how signalling cascades integrate with transcription factor circuits to govern the fine balance between TSC self-renewal and differentiation. In addition, breakthroughs in reprogramming strategies have enabled the generation of TSCs from fibroblasts. Here, 