Mouse-human neural crest chimeras: Not only a matter of black and white
Posted by MC, on 25 February 2016
The neural crest arises between neural and non-neural ectoderm and represents a somatic cell type with unique properties of multipotency. The neural crest cells (NCCs) migrate throughout the body and differentiate into a diverse array of cell types and tissues including the peripheral nervous system, enteric and sensory nervous system, Schwann cells, skin melanocytes, as well as connective tissues. The neural crest has major clinical relevance since it is disproportionately involved in both inherited and acquired developmental abnormalities termed neurocristopathies.
NCCs migration, development, and differentiation into various tissues have been studied in vivo in avian embryos, in studies pioneered by Nicole Le Douarin. Donor quail neural crest tissues were grafted into similar regions of stage-matched developing chick embryos, to generate quail-chick neural crest chimeras in ovo. The cells of the two species are easy to distinguish based on the ability to identify interphase nucleus of the quail, in which a large amount of heterochromatic DNA is found in the nucleoplasm, and different from that seen in chick where heterochromatin is dispersed within the nucleoplasm nucleolus. This chimeric system provided reliable information on the fate and ontogeny of the engrafted cells.
Few years later, Rudolf Jaenisch developed a mouse chimeric system that allows the assessment of the developmental potential and migration of mouse neural crest in vivo. Primary neural crest cells isolated from C57BL/6 mouse embryos and microinjected in utero into neurulating E8.5 albino BALB/c embryos were shown to contribute efficiently to pigmentation in the host animal. The resulting neural crest chimeras showed, however, different coat pigmentation contribution-patterns depending on the genotype of the host embryos. Whereas BALB/c neural crest chimeras showed limited donor cell pigment contribution, restricted largely to the head and limbs, KitW-sh/KitW-sh mutant mice used as hosts for chimeras, displayed extensive pigmentation throughout, often exceeding 50% of the coat. The KitW-sh/KitW-sh murine model carries an inversion that spans a 2.8 Mbp segment proximal to the c-Kit locus that disrupts its regulatory sequences and leads to a deficit of melanocytes, with no change in viability or fertility. This mouse line allows for an empty melanoblast niche in which the transplanted NCCs can incorporate without competition from the host endogenous cell populations. In contrast to BALB/c chimeras, where the donor melanoblasts appeared to have migrated primarily in the characteristic dorsoventral direction, in KitW-sh/KitW-sh mutants the injected cells appeared to migrate into the longitudinal direction as well, as if the cells were spreading through an empty niche. This is consistent with the absence of a functional endogenous melanoblast population in KitW-sh/KitW-sh mutants, in contrast to BALB/c mice, which contain a full complement of melanocytes.
The human induced pluripotent stem cell (hiPSCs) technology provides patient specific pluripotent cells that carry all genetic alterations that contributed to the disorder and thus represent a genetically defined cell system to study the respective disease. The greatest promise of hiPSCs is its potential to study human diseases in the Petri dish. In this approach patient-derived cells are differentiated into the cell types, which is affected in the patient with the goal to uncover a disease relevant phenotype in the dish. This method was also applied for modeling neurocristopathies such as Familial Dysautonomia, by differentiating patient-derived cells hiPSCs to neural crest and its derivatives, which presented disease manifestation in vitro. However, numerous human diseases originate already in embryogenesis, i.e. are caused by disturbances of developmental processes, therefore such an approach cannot recapitulate the developmental aspects of a disease because the test cells are not incorporated into the developing embryo and do not participate in normal developmental processes. Thus, a major challenge of the “disease in the dish” approach is establishing model systems that, using patient-derived hiPSCs, allow for the investigation of human disease under long-term in vivo conditions.
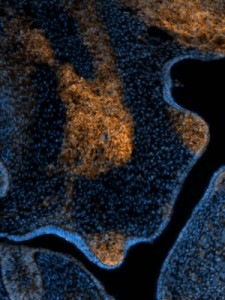
Inspired by the ability to generate neural crest chimeras, we aspired to use the human pluripotent stem cells as a cellular source of human NCCs and to generate mouse-human neural crest chimeras. This platform would expand the potential of using human pluripotent stem cells for studying human neural crest development and disease in vivo. In our recent paper we have shown that human NCCS derived from human pluripotent stem cells when microinjected into post-implantation KitW-sh/KitW-sh mouse embryos could participate in normal embryonic development and provide functional neural crest contribution to the host murine model. We tracked the implanted NCCs, which had been GFP labeled, thru their migration paths, and found that the human cells exhibited similar migration patterns as would normally be found in mice. As a result, about 30% of the implanted embryos showed human NCCs during development, and later in adult mice pigmentation, which is similar to what we found in mouse-mouse neural crest chimeras.
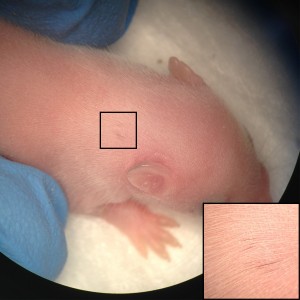
Our results are one of the first evidences of contribution of human embryonic cell population with functional evidence in adult postnatal mammal. Interestingly, we have observed that the largest the evolutionary distance between the NCCs and the host mice, the less the contribution is noticed. While a widespread pigmentation was observed using primary mouse NCCs as donor cells, only localized pigmented hairs were found in KitW-sh/KitW-sh host mice, using rat NCCs and minimal for in vitro derived human NCCs. The limited level of contribution we observed using human NCCs, presumably represents the over 90 million years evolutionary distance between mouse and human, suggesting that the host environment might limit the maturation and differentiation of the injected human NCCs.
Our work serves as proof of concept and an important first step toward the goal of generating chimeric mice that carry disease-relevant human cells in the relevant tissue. Resulting mouse-human chimeras would fill an important gap in disease research, as existing models do not accurately mimic certain diseases and disease states. Cancer is frequently studied using xenografts, however this approach fails to provide insight into tumor initiation and progression. Moreover, complex diseases with long latencies, such as Alzheimer’s and Parkinson’s disease, can only be partially modeled using induced pluripotent stem cells in vitro. Mouse-human chimeras would be used to overcome these limitations and could be used for regenerative medicine as well.


 (6 votes)
(6 votes)
 (4 votes)
(4 votes)