Here are the highlights from the current issue of Development:
Mammary gland RankL-ed into making milk
 Extensive remodelling of the mammary gland during pregnancy generates milk-producing lobuloalveolar structures. During remodelling, multipotent stem cells proliferate and differentiate into lineage-committed progenitor cells, which subsequently differentiate into mature epithelial cell types. It has been proposed that progesterone drives mammary stem cell expansion during pregnancy via paracrine signalling from progesterone receptor-positive sensor cells. On p. 1397, Christopher Ormandy and colleagues investigate how the resulting cells are directed towards the secretory lineage. Progesterone induces the expression of the transcription factor Elf5 in mouse mammary progenitor cells, they report, and Elf5 (which is essential for progenitor cell differentiation) cooperates with progesterone to promote alveolar development. They show that the progesterone receptor and Elf5 are expressed in different cell types, and identify the transcriptional activator RankL as the paracrine mediator of progesterone’s effects on Elf5 expression in progenitor cells and their consequent differentiation. RankL also mediates the progesterone-induced division of mammary stem cells. Together, these findings reveal how steroid hormones drive mammary gland remodelling during pregnancy.
Extensive remodelling of the mammary gland during pregnancy generates milk-producing lobuloalveolar structures. During remodelling, multipotent stem cells proliferate and differentiate into lineage-committed progenitor cells, which subsequently differentiate into mature epithelial cell types. It has been proposed that progesterone drives mammary stem cell expansion during pregnancy via paracrine signalling from progesterone receptor-positive sensor cells. On p. 1397, Christopher Ormandy and colleagues investigate how the resulting cells are directed towards the secretory lineage. Progesterone induces the expression of the transcription factor Elf5 in mouse mammary progenitor cells, they report, and Elf5 (which is essential for progenitor cell differentiation) cooperates with progesterone to promote alveolar development. They show that the progesterone receptor and Elf5 are expressed in different cell types, and identify the transcriptional activator RankL as the paracrine mediator of progesterone’s effects on Elf5 expression in progenitor cells and their consequent differentiation. RankL also mediates the progesterone-induced division of mammary stem cells. Together, these findings reveal how steroid hormones drive mammary gland remodelling during pregnancy.
Epidermal stem cell heterogeneity
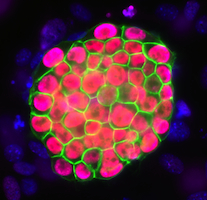
 Stochastic fluctuations in gene expression (transcriptional noise), which drive transitions between states, and complex regulatory networks, which define stable interconvertible ‘attractor’ states, are both thought to be involved in the control of stem cell fate. Because population-based studies lack the resolution to reveal the heterogeneity resulting from different attractor states or to uncover their functional significance, on p. 1433 Fiona Watt and colleagues use single-cell gene expression profiling to investigate heterogeneity among human epidermal stem cells. Markers for these cells include the Notch ligand delta-like 1 (DLL1) and the EGFR antagonist LRIG1, but the expression of these proteins is known to vary between cells. The researchers now show that DLL1 expression discriminates between two populations of undifferentiated basal keratinocytes that differ in adhesion and proliferation, and that have distinct transcriptional profiles. These results indicate that intercellular differences in gene expression among human epidermal stem cells cannot be attributed solely to random stochastic fluctuations and suggest that this heterogeneity affects the interaction of these cells with their environment.
Stochastic fluctuations in gene expression (transcriptional noise), which drive transitions between states, and complex regulatory networks, which define stable interconvertible ‘attractor’ states, are both thought to be involved in the control of stem cell fate. Because population-based studies lack the resolution to reveal the heterogeneity resulting from different attractor states or to uncover their functional significance, on p. 1433 Fiona Watt and colleagues use single-cell gene expression profiling to investigate heterogeneity among human epidermal stem cells. Markers for these cells include the Notch ligand delta-like 1 (DLL1) and the EGFR antagonist LRIG1, but the expression of these proteins is known to vary between cells. The researchers now show that DLL1 expression discriminates between two populations of undifferentiated basal keratinocytes that differ in adhesion and proliferation, and that have distinct transcriptional profiles. These results indicate that intercellular differences in gene expression among human epidermal stem cells cannot be attributed solely to random stochastic fluctuations and suggest that this heterogeneity affects the interaction of these cells with their environment.
Adipocytes and wound healing
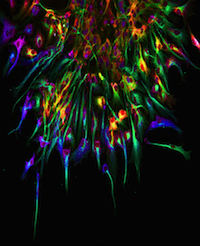
 Acute wound healing, which restores the essential barrier function of the skin, requires the coordination of the proliferation and migration of both keratinocytes and fibroblasts for epidermal and dermal repair, respectively. Skin healing is known to involve communication between haematopoietic cells, keratinocytes and fibroblasts, but could it involve other intradermal cell types? Barbara Schmidt and Valerie Horsley (p. 1517) have been investigating the involvement of intradermal adipocytes in skin wound healing in mice. They report that immature adipocytes are activated during the proliferative phase of acute skin wound healing and that mature adipocytes appear in healing wounds concurrently with fibroblasts. Moreover, by studying wound healing in mice with genetic defects in adipogenesis and in mice treated with pharmacological inhibitors of adipogenesis, they provide evidence that suggests that adipocytes are necessary for fibroblast recruitment and for dermal reconstruction. Further experiments are now needed to elucidate how adipocytes mediate fibroblast function during the healing of acute skin wounds and to determine whether they also aid the healing of chronic wounds.
Acute wound healing, which restores the essential barrier function of the skin, requires the coordination of the proliferation and migration of both keratinocytes and fibroblasts for epidermal and dermal repair, respectively. Skin healing is known to involve communication between haematopoietic cells, keratinocytes and fibroblasts, but could it involve other intradermal cell types? Barbara Schmidt and Valerie Horsley (p. 1517) have been investigating the involvement of intradermal adipocytes in skin wound healing in mice. They report that immature adipocytes are activated during the proliferative phase of acute skin wound healing and that mature adipocytes appear in healing wounds concurrently with fibroblasts. Moreover, by studying wound healing in mice with genetic defects in adipogenesis and in mice treated with pharmacological inhibitors of adipogenesis, they provide evidence that suggests that adipocytes are necessary for fibroblast recruitment and for dermal reconstruction. Further experiments are now needed to elucidate how adipocytes mediate fibroblast function during the healing of acute skin wounds and to determine whether they also aid the healing of chronic wounds.
Blood vessels guide heart innervation
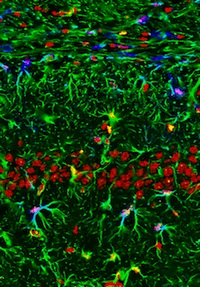
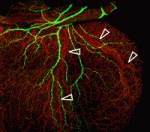 Peripheral nerves and blood vessels lie close together and have similar branching patterns in many tissues, which suggests that their development might be coordinated. Previously, Yoh-suke Mukouyama and co-workers reported that signals secreted by sensory nerves determine vascular patterning in the skin. Here (p. 1475), the researchers show that there is an anatomical congruence between sympathetic nerves and coronary veins in the developing mouse heart. In contrast to the situation in skin, this neurovascular association is not important for vascular patterning, but is instead required for the functional sympathetic innervation of the heart. Vascular smooth muscle cells (VSMCs) associate with coronary veins during angiogenic remodelling, they report, and mediate the extension of sympathetic axons within the subepicardium by secreting nerve growth factor (NGF). Subsequently, venous VSMCs downregulate NGF expression and arterial VSMCs begin to secrete NGF, which stimulates the axons to innervate coronary arteries in the myocardium. Thus, sequential expression of NGF in subepicardial venous VSMCs and myocardial arterial VSMCs determines the heart’s stereotypical pattern of sympathetic innervation.
Peripheral nerves and blood vessels lie close together and have similar branching patterns in many tissues, which suggests that their development might be coordinated. Previously, Yoh-suke Mukouyama and co-workers reported that signals secreted by sensory nerves determine vascular patterning in the skin. Here (p. 1475), the researchers show that there is an anatomical congruence between sympathetic nerves and coronary veins in the developing mouse heart. In contrast to the situation in skin, this neurovascular association is not important for vascular patterning, but is instead required for the functional sympathetic innervation of the heart. Vascular smooth muscle cells (VSMCs) associate with coronary veins during angiogenic remodelling, they report, and mediate the extension of sympathetic axons within the subepicardium by secreting nerve growth factor (NGF). Subsequently, venous VSMCs downregulate NGF expression and arterial VSMCs begin to secrete NGF, which stimulates the axons to innervate coronary arteries in the myocardium. Thus, sequential expression of NGF in subepicardial venous VSMCs and myocardial arterial VSMCs determines the heart’s stereotypical pattern of sympathetic innervation.
Timely BMP patterns neural tube
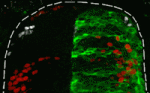 In many embryonic tissues, graded signals (morphogens) provide the positional information that governs the pattern of cellular differentiation. It is widely thought that cells interpret morphogen gradients by producing corresponding levels of intracellular signalling activity, which regulate differential gene expression. But could other, distinct, mechanisms be used to interpret some morphogens? On p. 1467, James Briscoe and co-workers investigate the morphogen activity of bone morphogenetic protein (BMP) signalling in the chick dorsal neural tube. BMPs are thought to provide the positional information that specifies the spatial pattern of the dorsal interneurons. In neural tube explants, the researchers report, the duration of exposure to Bmp4 generates distinct levels of intracellular signalling and induces specific dorsal identities. Moreover, they find that a dynamic gradient of BMP activity progressively specifies more dorsal identities in the neural tube in vivo. Given these results, the researchers propose a model for morphogen interpretation in which the temporally dynamic control of signalling is required to specify the spatial pattern of cellular differentiation.
In many embryonic tissues, graded signals (morphogens) provide the positional information that governs the pattern of cellular differentiation. It is widely thought that cells interpret morphogen gradients by producing corresponding levels of intracellular signalling activity, which regulate differential gene expression. But could other, distinct, mechanisms be used to interpret some morphogens? On p. 1467, James Briscoe and co-workers investigate the morphogen activity of bone morphogenetic protein (BMP) signalling in the chick dorsal neural tube. BMPs are thought to provide the positional information that specifies the spatial pattern of the dorsal interneurons. In neural tube explants, the researchers report, the duration of exposure to Bmp4 generates distinct levels of intracellular signalling and induces specific dorsal identities. Moreover, they find that a dynamic gradient of BMP activity progressively specifies more dorsal identities in the neural tube in vivo. Given these results, the researchers propose a model for morphogen interpretation in which the temporally dynamic control of signalling is required to specify the spatial pattern of cellular differentiation.
Grandmother, what big teeth you have!
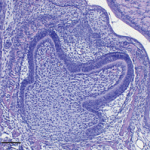 Although many organs renew throughout life, the regenerative capacity of some organs varies between species. For example, reptiles replace their teeth continuously, whereas mammals have only a single round of tooth replacement. The development of new teeth involves dental epithelial cells that are competent for tooth formation. Now, Irma Thesleff and colleagues (p. 1424) identify the transcription factor Sox2 as a marker for these cells. The researchers report that Sox2 is expressed in the dental lamina that gives rise to successional teeth in ferrets and humans, which have a single round of tooth replacement, and in five reptiles with continuous tooth replacement. Sox2 is also expressed in the dental lamina during the serial addition of mammalian molars and, in mice, Sox2+ cells in the first molar give rise to the sequentially developing second and third molars. These data suggest that Sox2 function has been conserved during evolution, and identify tooth replacement and the serial addition of primary teeth as variations of the same developmental process.
Although many organs renew throughout life, the regenerative capacity of some organs varies between species. For example, reptiles replace their teeth continuously, whereas mammals have only a single round of tooth replacement. The development of new teeth involves dental epithelial cells that are competent for tooth formation. Now, Irma Thesleff and colleagues (p. 1424) identify the transcription factor Sox2 as a marker for these cells. The researchers report that Sox2 is expressed in the dental lamina that gives rise to successional teeth in ferrets and humans, which have a single round of tooth replacement, and in five reptiles with continuous tooth replacement. Sox2 is also expressed in the dental lamina during the serial addition of mammalian molars and, in mice, Sox2+ cells in the first molar give rise to the sequentially developing second and third molars. These data suggest that Sox2 function has been conserved during evolution, and identify tooth replacement and the serial addition of primary teeth as variations of the same developmental process.
PLUS…
Crossing paths: cytokinin signalling and crosstalk
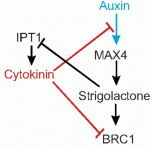 Cytokinins are a major class of plant hormones that are involved in various aspects of plant development. Recent studies, reviewed here by Helariutta and colleagues, have shown that cytokinins interact with a number of other plant hormones to regulate plant development.
Cytokinins are a major class of plant hormones that are involved in various aspects of plant development. Recent studies, reviewed here by Helariutta and colleagues, have shown that cytokinins interact with a number of other plant hormones to regulate plant development.
See the Review article on p. 1373
Conserved non-coding elements and cis regulation: actions speak louder than words
 The literature is strewn with examples of conserved non-coding elements being able to drive reporter expression, but the extent to which such sequences are actually used endogenously in vivo has been unclear. Here, Nelson and Wardle outline the various methods used to define cis-regulatory elements driving gene expression, and discuss the complex relationship between sequence conservation and the functional equivalence of such elements during development and across evolution.
The literature is strewn with examples of conserved non-coding elements being able to drive reporter expression, but the extent to which such sequences are actually used endogenously in vivo has been unclear. Here, Nelson and Wardle outline the various methods used to define cis-regulatory elements driving gene expression, and discuss the complex relationship between sequence conservation and the functional equivalence of such elements during development and across evolution.
See the Review article on p. 1385
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (3 votes)
(3 votes)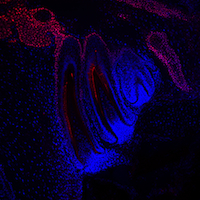



 Extensive remodelling of the mammary gland during pregnancy generates milk-producing lobuloalveolar structures. During remodelling, multipotent stem cells proliferate and differentiate into lineage-committed progenitor cells, which subsequently differentiate into mature epithelial cell types. It has been proposed that progesterone drives mammary stem cell expansion during pregnancy via paracrine signalling from progesterone receptor-positive sensor cells. On
Extensive remodelling of the mammary gland during pregnancy generates milk-producing lobuloalveolar structures. During remodelling, multipotent stem cells proliferate and differentiate into lineage-committed progenitor cells, which subsequently differentiate into mature epithelial cell types. It has been proposed that progesterone drives mammary stem cell expansion during pregnancy via paracrine signalling from progesterone receptor-positive sensor cells. On  Stochastic fluctuations in gene expression (transcriptional noise), which drive transitions between states, and complex regulatory networks, which define stable interconvertible ‘attractor’ states, are both thought to be involved in the control of stem cell fate. Because population-based studies lack the resolution to reveal the heterogeneity resulting from different attractor states or to uncover their functional significance, on
Stochastic fluctuations in gene expression (transcriptional noise), which drive transitions between states, and complex regulatory networks, which define stable interconvertible ‘attractor’ states, are both thought to be involved in the control of stem cell fate. Because population-based studies lack the resolution to reveal the heterogeneity resulting from different attractor states or to uncover their functional significance, on  Acute wound healing, which restores the essential barrier function of the skin, requires the coordination of the proliferation and migration of both keratinocytes and fibroblasts for epidermal and dermal repair, respectively. Skin healing is known to involve communication between haematopoietic cells, keratinocytes and fibroblasts, but could it involve other intradermal cell types? Barbara Schmidt and Valerie Horsley (
Acute wound healing, which restores the essential barrier function of the skin, requires the coordination of the proliferation and migration of both keratinocytes and fibroblasts for epidermal and dermal repair, respectively. Skin healing is known to involve communication between haematopoietic cells, keratinocytes and fibroblasts, but could it involve other intradermal cell types? Barbara Schmidt and Valerie Horsley ( Peripheral nerves and blood vessels lie close together and have similar branching patterns in many tissues, which suggests that their development might be coordinated. Previously, Yoh-suke Mukouyama and co-workers reported that signals secreted by sensory nerves determine vascular patterning in the skin. Here (
Peripheral nerves and blood vessels lie close together and have similar branching patterns in many tissues, which suggests that their development might be coordinated. Previously, Yoh-suke Mukouyama and co-workers reported that signals secreted by sensory nerves determine vascular patterning in the skin. Here ( In many embryonic tissues, graded signals (morphogens) provide the positional information that governs the pattern of cellular differentiation. It is widely thought that cells interpret morphogen gradients by producing corresponding levels of intracellular signalling activity, which regulate differential gene expression. But could other, distinct, mechanisms be used to interpret some morphogens? On
In many embryonic tissues, graded signals (morphogens) provide the positional information that governs the pattern of cellular differentiation. It is widely thought that cells interpret morphogen gradients by producing corresponding levels of intracellular signalling activity, which regulate differential gene expression. But could other, distinct, mechanisms be used to interpret some morphogens? On  Although many organs renew throughout life, the regenerative capacity of some organs varies between species. For example, reptiles replace their teeth continuously, whereas mammals have only a single round of tooth replacement. The development of new teeth involves dental epithelial cells that are competent for tooth formation. Now, Irma Thesleff and colleagues (
Although many organs renew throughout life, the regenerative capacity of some organs varies between species. For example, reptiles replace their teeth continuously, whereas mammals have only a single round of tooth replacement. The development of new teeth involves dental epithelial cells that are competent for tooth formation. Now, Irma Thesleff and colleagues ( Cytokinins are a major class of plant hormones that are involved in various aspects of plant development. Recent studies, reviewed here by Helariutta and colleagues, have shown that cytokinins interact with a number of other plant hormones to regulate plant development.
Cytokinins are a major class of plant hormones that are involved in various aspects of plant development. Recent studies, reviewed here by Helariutta and colleagues, have shown that cytokinins interact with a number of other plant hormones to regulate plant development. The literature is strewn with examples of conserved non-coding elements being able to drive reporter expression, but the extent to which such sequences are actually used endogenously in vivo has been unclear. Here, Nelson and Wardle outline the various methods used to define cis-regulatory elements driving gene expression, and discuss the complex relationship between sequence conservation and the functional equivalence of such elements during development and across evolution.
The literature is strewn with examples of conserved non-coding elements being able to drive reporter expression, but the extent to which such sequences are actually used endogenously in vivo has been unclear. Here, Nelson and Wardle outline the various methods used to define cis-regulatory elements driving gene expression, and discuss the complex relationship between sequence conservation and the functional equivalence of such elements during development and across evolution.