In Development this week (Vol. 138, Issue 24)
Posted by Seema Grewal, on 22 November 2011
Here are the research highlights from the current issue of Development:
Getting to the heart of Flk1 expression
The Flk1 gene, which encodes a VEGF-A receptor, is expressed in the multipotent mesodermal progenitor cells of mouse embryos that give rise to various haemato-cardiovascular cell lineages. FLK1 expression also marks haemato-cardiovascular cell lineages in differentiating human embryonic stem (ES) cells, lineages that could be useful for the treatment of human cardiovascular diseases if the molecular regulation of Flk1 expression can be unravelled. Masatsugu Ema and colleagues now identify a novel enhancer in the mouse Flk1 gene that is required for the mesodermal expression of Flk1 in the early embryo and in differentiating ES cells (see p. 5357). This enhancer region, they report, is activated by Bmp, Wnt and Fgf, and contains binding sites for the transcription factors Gata, Cdx, Tcf/Lef, ER71/Etv2 and Fox, all of which are controlled by Bmp, Wnt and Fgf signalling. The researchers suggest, therefore, that early Flk1 expression might be induced by cooperative interactions between this set of transcription factors under the control of Bmp, Wnt and Fgf signalling.
Eph/ephrin signals guide muscle rebuilding
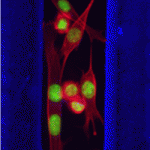
Skeletal muscle regeneration after injury is dependent on satellite cells (skeletal muscle stem cells) that, in response to local myofibre damage, proliferate to build up a supply of adult myoblasts that repair the damage. But do satellite cells relocate within the muscle to respond to distant myofibre damage? If so, how do they find their way? On p. 5279, D. D. W. Cornelison and co-workers investigate whether Ephs and ephrins – molecules that are usually associated with axon guidance but that are expressed by activated satellite cells – modulate satellite cell motility and patterning. Using an ephrin ‘stripe’ assay, they show that multiple ephrins elicit a repulsive migratory response in activated satellite cells and affect the patterning of differentiating satellite cells. Importantly, the same ephrins are present on the surface of healthy myofibres and increase during regeneration, which suggests that muscle regeneration could involve ephrin-mediated guidance. Given their results, the researchers propose that Eph/ephrin signalling might regulate multiple aspects of satellite cell behaviour during muscle regeneration.
Noncanonical Wnts and PAR-1 drive neural crest fate
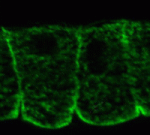
Neural crest (NC) cells are multipotent progenitors that form at the neural plate border, undergo epithelial-mesenchymal transition, and then migrate to give rise to numerous cell types in vertebrate embryos. Noncanonical Wnt signalling is known to be involved in NC migration, but is it, like canonical Wnt signalling, required for NC specification? On p. 5441, Olga Ossipova and Sergei Sokol implicate noncanonical Wnt11-like proteins in NC specification in Xenopus embryos. They show that Wnt11R, which is expressed in the neuroectoderm next to the NC territory, is required for NC formation. The authors also show that Wnt11-like signals regulate the localisation and activity of the cell polarity determinant PAR-1. Importantly, PAR-1 itself is required for NC specification, they report, and PAR-1 RNA rescues NC markers in embryos in which noncanonical Wnt signalling has been blocked. Together, these results identify roles for noncanonical Wnt signalling and PAR-1 in NC specification and reveal an unexpected connection between cell polarisation and cell fate.
Dual embryonic origin for the inner ear
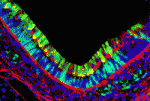
It is widely accepted that the inner ear labyrinth and the neurons of the cochleovestibular ganglion (CVG), which innervates the inner ear’s sensory epithelia, derive entirely from the otic placode, an ectodermal region that invaginates during embryogenesis to form the otic vesicle (OV) and the CVG. Here (p. 5403), by genetically labelling cranial neuroepithelial cell (NEC) lineages, including neural crest cells, in mice, Bernice Morrow and colleagues show that cells from the neural tube invade the otic epithelium in vivo and that NEC descendants constitute a significant proportion of the OV. NEC descendants, they report, are localised within the sensory epithelia of the saccule and utricle (the inner ear structures that are sensitive to movement) and the cochlea (the auditory portion of the inner ear) throughout development and into adulthood, and differentiate into neurons, hair cells and supporting cells. By revealing the inner ear’s dual embryonic origin, these results challenge the current model for the neurosensory development of the inner ear.
Heads up for new Noggin functions
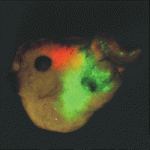
The secreted protein Noggin1 antagonises the BMP family of TGFβ ligands and, as a consequence, plays a key role in many processes during embryogenesis. Here (p. 5345), Andrey Zaraisky and colleagues unexpectedly reveal that Noggin1 and its homologue Noggin2 also antagonise, albeit less effectively, the non-BMP TGFβ ligands ActivinB, Xnr2 and Xnr4 (Nodal homologues), and XWnt8 during early Xenopus embryogenesis. Inactivation of these ligands is essential for head induction, and the researchers show that both Noggin proteins can induce a secondary head, including a forebrain, if ectopically produced at high concentrations in Xenopus embryos. During normal development, they report, the Noggin1 concentration in the presumptive forebrain is only sufficient for its BMP-antagonizing function whereas the higher concentration of Noggin2 produced in the anterior margin of the neural plate protects the developing forebrain from inhibition by ActivinB and XWnt8 signalling. Thus, the researchers conclude, forebrain specification in Xenopus requires the inhibition of Activin/Nodal, BMP and Wnt signalling not only during gastrulation but also at post-gastrulation stages.
Seven up works double time in neuroblasts

Neural progenitor cells generate different cell types at different times during nervous system development. In Drosophila neuroblasts, the sequential expression of Hunchback (Hb), Kruppel (Kr) and several other transcription factors controls temporal competence changes. The transcription factors in this temporal cascade regulate each other’s expression but, in addition, Seven up (Svp) acts as a switching factor to ensure the Hb to Kr transition. Now, Stefan Thor and co-workers uncover a second role for Svp during the development of the Drosophila embryonic thoracic neuroblast 5-6 (NB5-6T) lineage (see p. 5311). The researchers show that svp is expressed in two distinct pulses in this lineage. In the early pulse, they report, svp acts as a switching factor by suppressing hb expression. However, in the second pulse, which occurs later in the NB5-6T lineage, svp acts as a sub-temporal gene to establish the alternative fates of four interneurons expressing the transcription factor Apterous. Thus, one gene can play two temporal roles in the development of one neural lineage.
Plus…
This issue, the last of the 2011 volume, contains Development’s annual Book Review section, which covers a broad range of topics that are becoming increasingly important to developmental biologists. The titles reviewed include:
– Mathematical Models of Biological Systems
(reviewed by Lance Davidson)
– Principles of Development
(reviewed by Richard Harland)
– Imaging in Developmental Biology: A Laboratory Manual
(reviewed by Elaine Dzierzak and Catherine Robin)
– Molecular Biology of RNA
(reviewed by Ilan Davis)
– Epigenetics Linking Genotype and Phenotype in Development and Evolution
(reviewed by Mellissa R. W. Mann)
– The Nucleus
(reviewed by Wendy A. Bickmore)
– Human Stem Cell Technology and Biology: A Research Guide and Laboratory Manual
(reviewed by Neil Singh and Ludovic Vallier)


 (No Ratings Yet)
(No Ratings Yet) (3 votes)
(3 votes)