Behind the paper: Uncovering the dual origins of human cortical astrocytes
Posted by Denise Allen, on 18 August 2022
Dr. Denise Allen and Dr. Tomasz Nowakowski at the University of California, San Francisco recently published an article in Science where they revealed a dual origin for astrocytes in the human cortex. Using a combination of fate mapping and single cell analysis, they revealed that the two stem cell niches in the developing cortex give rise to spatially, morphologically, and molecularly distinct populations of astrocytes. The Node asked them to give us a behind the scenes look at how the story came together:
- How did you get started on this project?
TJN: For a very long time, we have been interested in the question of why the brains of primates and humans are so much larger and complex than the brains of mice, which we study frequently in the laboratory. Differences in brain size can be found very early on during development, and therefore it was plausible to hypothesize that differences in the way radial glia, which act as neural stem cells, develop could contribute to these differences. In our prior work, we found that animals with large brains, such as primates or humans, contain a greater diversity of radial glia subtypes compared to mice. In particular, we found that based on gene expression profiles, radial glia could be divided into truncated radial glia and outer radial glia, which are located in two anatomically distinct niches of the developing cortex.
DA: During my undergraduate neuroscience classes I was always struck by the deep knowledge we have about the development of neurons in the cerebral cortex, but astrocytes and other glia seemed to be so often overlooked. During my rotation in the Nowakowski lab, I became fascinated with Tom’s preliminary data that suggested distinct subtypes of radial glia could give rise to distinct astrocyte populations. I was really excited by the fact that large brain mammals, including primates and humans, seem to have a different repertoire of radial glia compared to rodents, as well as much more complex astrocytes. So the possibility to study the unique features of human development with a focus on astrocytes was a dream come true.
- What was already known about the developmental trajectories of radial glia in the developing brain prior to your work?
A lot of work has been done probing the differentiation of outer radial glia (also known as basal radial glia). Numerous papers have shown that they give rise to neurons, oligodendrocytes and supposedly the majority of astrocytes, but the role of truncated radial glia has not been studied in great detail. Previous studies have suggested that because few mitotic cells can be found in the ventricular zone stem cell niche during midgestation in primates and humans, that the truncated radial glia that reside in this zone are unlikely to serve as a major source of new cells. We decided to challenge this assumption by labeling progenitors in the ventricular zone and determining the fates of the resulting cells. To our surprise, we found that neurons, oligodendrocytes, and astrocytes continue to be produced by ventricular zone progenitors.
- Can you summarize your findings? What was the key experiment?
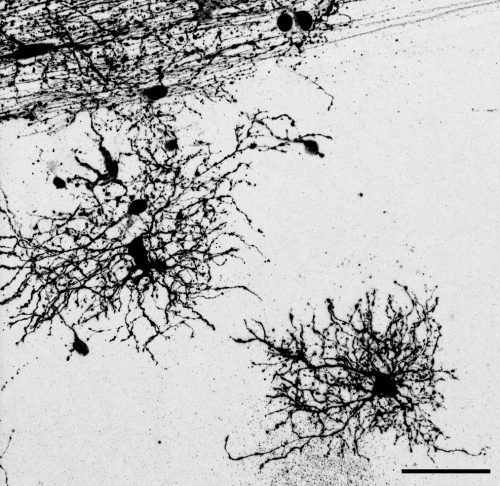
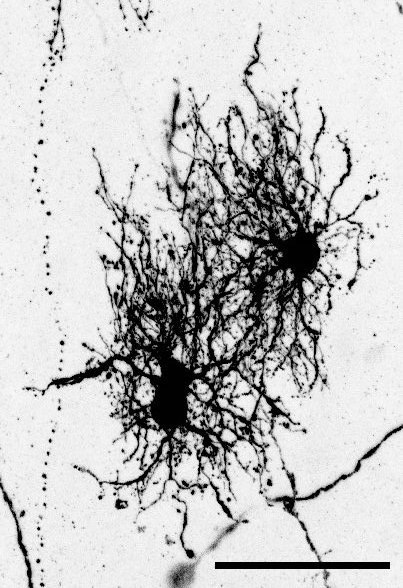
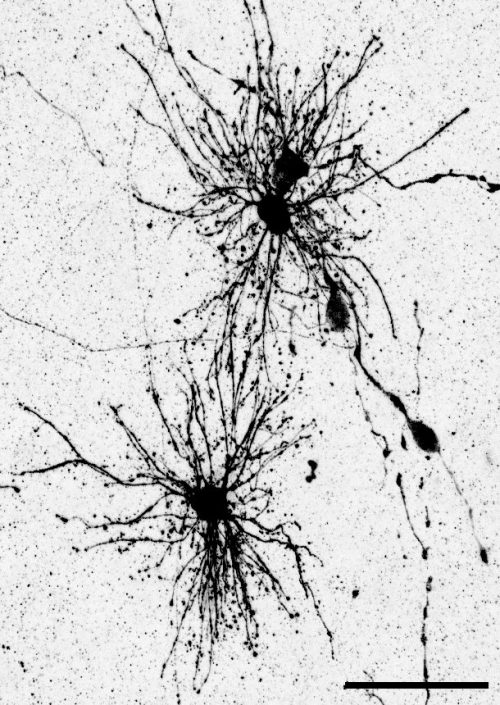
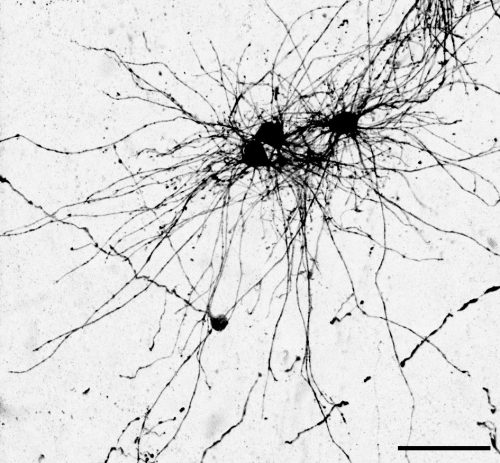
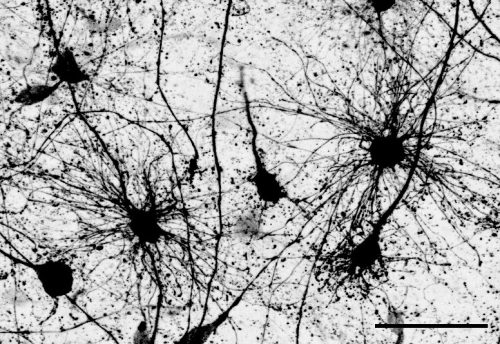
The key experiment involved labeling progenitor cells that occupy anatomically distinct niches that truncated and outer radial glia cells, and tracing the fates of cells that they produce. We found that while both populations broadly produce similar cell types (neurons, astrocytes and oligodendrocytes), they produce very distinct subtypes of astrocytes. In a series of very striking results, we found that truncated radial glia give rise to astrocytes that migrate to the cortical plate, while outer radial glia give rise to astrocytes that do not migrate, and instead differentiate locally in the outer subventricular zone.
We often speak about the diversity of neurons, but classical studies have also shown that astrocytes can be remarkably diverse, even in early development. I wanted to further explore the diversity of the astrocyte subtypes we identified, but it is challenging to connect these classical descriptions of astrocyte subtypes to modern-day descriptions such as those derived from single cell sequencing. To solve this problem, I took advantage of a method called Patch-seq which gave us the ability to aspirate the contents of a cell that was previously defined based on its morphology and position, and then performing sequencing of that cellular contents to determine a molecular identity. This analysis was key for bridging our cellular definitions based on morphology and developmental cell lineage, and linking them to molecular markers. This allowed me to bring the story in full circle.
- When doing the research, did you have any particular result or eureka moment that has stuck with you?
The very first experiment I performed that involved labeling these two different stem cell niches resulted in a distribution of cell types that could not have been more different. Many comparisons in developmental biology rest upon small differences between conditions. To see such a stark difference in the distribution of cells–especially of glia–was an exciting moment that defined the course of the project very early on in my PhD.
Another surprising finding was when we started closely comparing classical drawings by Ramon y Cajal and Retzius and to images of our astrocyte subtypes. Remarkably, we found that our “dense bulbous” astrocytes were clearly depicted in those early records, but these cells have rarely been mentioned in modern literature. This realization gave us a lot of confidence that the cells we were observing were a real phenomenon and not an experimental artifact. These cells had just been lying in wait, waiting for someone to put the spotlight on them.
- And what about the flipside? Any frustrations or despair?
The Universe really conspired against us when we were trying to finish experiments for the revision of the paper. I set up the last three revision experiments in late December, when we suddenly found out that it was time to move our lab to a new building at UCSF. We came up with an elaborate system to keep the cultures going while we moved and they seemed to have survived, until someone suddenly noticed that the incubator had failed and the alarm hadn’t gone off. What followed was two months of issue after issue trying to repeat these last two experiments, but we finally got there in the end!
- Where will this story take the lab?
This work has inspired several new projects in the lab. We are excited to examine if similar findings can be replicated in other models of brain development such as cerebral organoids, what these unique subpopulations look like in the adult brain, and what role they might play in disease states. I’m also hoping this work will also attract more trainees interested in glial development to the lab!
- What is next for you/the lab after this paper? Let us know if you are continuing this research, or starting/looking for a new position.
Denise has graduated and is currently interviewing for computational biologist roles in biotech. She is looking forward to delving into the “big data” side of biology, and working towards making a significant impact on patients’ lives.


 (1 votes)
(1 votes)