June in preprints
Posted by the Node, on 1 July 2020
Welcome to our monthly trawl for developmental biology (and related) preprints.
Preprints hosted on bioRxiv and arXiv, use these links to get to the section you want.
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice & education
Developmental biology
| Patterning & signalling
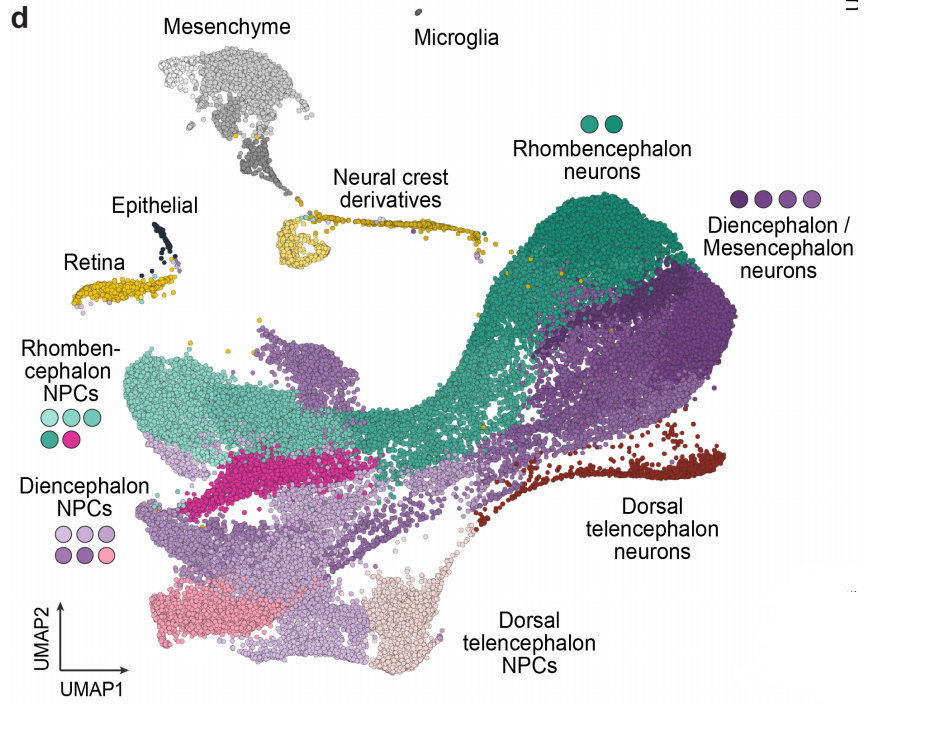
Lineage recording reveals dynamics of cerebral organoid regionalization
Zhisong He, Tobias Gerber, Ashley Maynard, Akanksha Jain, Rebecca Petri, Malgorzata Santel, Kevin Ly, Leila Sidow, Fatima Sanchis Calleja, Stephan Riesenberg, J. Gray Camp, Barbara Treutlein
VEGFC induced cell cycle arrest mediates sprouting and differentiation of venous and lymphatic endothelial cells
Ayelet Jerafi-Vider, Noga Moshe, Gideon Hen, Daniel Splittstoesser, Masahiro Shin, Nathan Lawson, Karina Yaniv
Akt is required for artery formation during embryonic vascular development
Wenping Zhou, Emma Ristori, Liqun He, Joey J Ghersi, Sameet Mehta, Rong Zhang, Christer Betsholtz, Stefania Nicoli, William C. Sessa
Remodeling mechanisms determine size distributions in developing retinal vasculature
Osamu Iizuka, Shotaro Kawamura, Atsushi Tero, Akiyoshi Uemura, Takashi Miura
Environmental Oxygen Regulates Astrocyte Proliferation to Guide Angiogenesis during Retinal Development
Robin M Perelli, Matthew L O’Sullivan, Samantha Zarnick, Jeremy N Kay
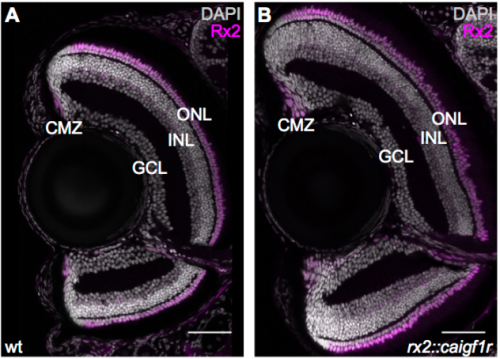
Igf signalling uncouples retina growth from body size by modulating progenitor cell division
Clara Becker, Katharina Lust, Joachim Wittbrodt
Axial skeleton anterior-posterior patterning is regulated through feedback regulation between Meis transcription factors and retinoic acid
Alejandra C. López-Delgado, Irene Delgado, Vanessa Cadenas, Fátima Sánchez-Cabo, Miguel Torres
Defining the signalling determinants of a posterior ventral spinal cord identity in human neuromesodermal progenitor derivatives
Matthew Wind, Antigoni Gogolou, Ichcha Manipur, Ilaria Granata, Larissa Butler, Peter W. Andrews, Ivana Barbaric, Ke Ning, Mario R. Guarracino, Marysia Placzek, Anestis Tsakiridis
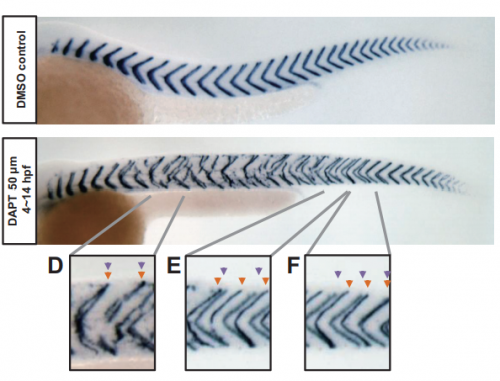
From local resynchronization to global pattern recovery in the zebrafish segmentation clock
Koichiro Uriu, Bo-Kai Liao, Andrew C. Oates, Luis G. Morelli
Evidence of progenitor cell lineage rerouting in the adult mouse hippocampus
Daniela M.S. Moura, Juliana Alves Brandão, Celia Lentini, Christophe Heinrich, Claudio M. Queiroz, Marcos R. Costa
Cascade Diversification Directs the Generation of Neuronal Diversity in Hypothalamus
Yu-Hong Zhang, Mingrui Xu, Si Li, Haoda Wu, Xiang Shi, Xize Guo, Wenhui Mu, Ling Gong, Mingze Yao, Miao He, Qing-Feng Wu
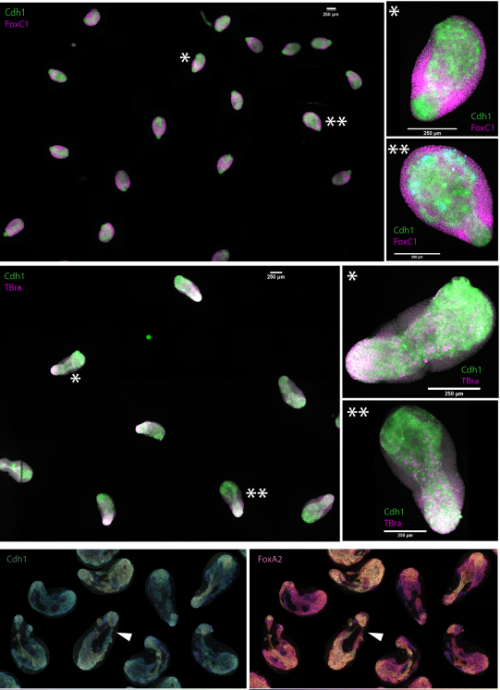
Cell-state transitions and collective cell movement generate an endoderm-like region in gastruloids
Ali Hashmi, Sham Tlili, Pierre Perrin, Alfonso Martinez-Arias, Pierre-François Lenne
In vitro endoderm emergence and self-organisation in the absence of extraembryonic tissues and embryonic architecture
Stefano Vianello, Matthias P. Lutolf
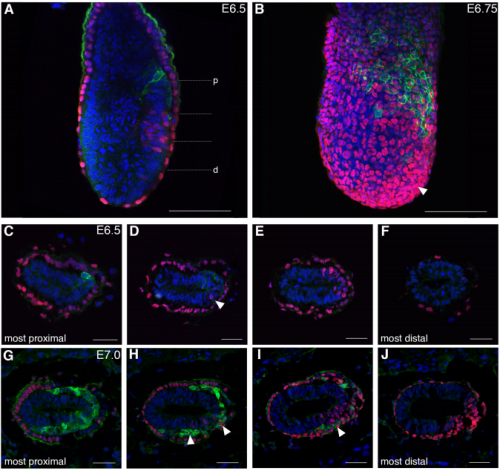
Spatiotemporal sequence of mesoderm and endoderm lineage segregation during mouse gastrulation
Simone Probst, Sagar, Jelena Tosic, Carsten Schwan, Dominic Grün, Sebastian J. Arnold
Rostrocaudal Patterning and Neural Crest Differentiation of Human Pre-Neural Spinal Cord Progenitors in vitro
Fay Cooper, George E Gentsch, Richard Mitter, Camille Bouissou, Lyn Healy, Ana Hernandez-Rodriguez, James C Smith, Andreia S Bernardo
De novo enteric neurogenesis in post-embryonic zebrafish from Schwann cell precursors rather than resident cell types
Wael Noor El-Nachef, Marianne E. Bronner
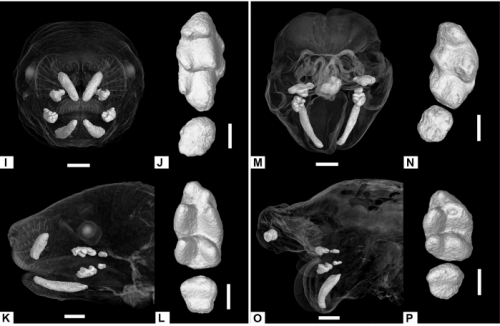
Anomalous incisor morphology indicates tissue-specific roles for Tfap2a and Tfap2b in tooth development
Emily D. Woodruff, Galaxy C. Gutierrez, Eric Van Otterloo, Trevor Williams, Martin J. Cohn
Constitutional activation of BMP4 and WNT signalling in hESC results in impaired mesendoderm differentiation
C. Markouli, E. Couvreu De Deckersberg, D. Dziedzicka, M. Regin, S. Franck, A. Keller, A. Gheldof, M. Geens, K. Sermon, C. Spits
Stabilization of β-catenin promotes melanocyte specification at the expense of the Schwann cell lineage
Sophie Colombo, Valérie Petit, Roselyne Y Wagner, Delphine Champeval, Ichiro Yajima, Franck Gesbert, Irwin Davidson, Veronique Delmas, Lionel Larue
Hnf4a is required for the development of Cdh6-expressing progenitors into proximal tubules in the mouse kidney
Sierra S. Marable, Eunah Chung, Joo-Seop Park
APP binds to the EGFR ligands HB-EGF and EGF, acting synergistically with EGF to promote ERK signaling and neuritogenesis
Joana F. da Rocha, Luísa Bastos, Sara C. Domingues, Ana R. Bento, Uwe Konietzko, Odete A. B. da Cruz e Silva, Sandra I. Vieira
Precise levels of Nectin-3 and an interaction with Afadin are required for proper synapse formation in postnatal visual cortex
Johanna Tomorsky, Philip R. L. Parker, Chris Q. Doe, Cristopher M. Niell
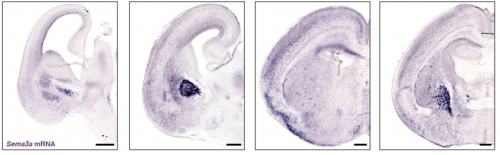
PlexinA4-Semaphorin3A mediated crosstalk between main cortical interneuron classes is required for superficial interneurons lamination
Greta Limoni, Mathieu Niquille, Sahana Murthy, Denis Jabaudon, Alexandre Dayer
Population dynamics and neuronal polyploidy in the developing neocortex
Thomas Jungas, Mathieu Joseph, Mohamad-Ali Fawal, Alice Davy
EXOC1 regulates cell morphology of spermatogonia and spermatocytes in mice
Yuki Osawa, Miho Usui, Yumeno Kuba, Hoai Thu Le, Natsuki Mikami, Toshinori Nakagawa, Yoko Daitoku, Kanako Kato, Hossam Hassan Shawki, Yoshihisa Ikeda, Akihiro Kuno, Kento Morimoto, Yoko Tanimoto, Tra Thi Huong Dinh, Kazuya Murata, Ken-ichi Yagami, Masatsugu Ema, Shosei Yoshida, Satoru Takahashi, Seiya Mizuno, Fumihiro Sugiyama
HES1 is a Critical Mediator of the SHH-GLI3 Axis in Regulating Digit Number
Deepika Sharma, Anthony J. Mirando, Abigail Leinroth, Jason T. Long, Courtney M. Karner, Matthew J. Hilton
A subpopulation of astrocyte progenitors defined by Sonic hedgehog signaling
Ellen Gingrich, Kendra Case, A. Denise R. Garcia
NMDA receptors control cortical axonal projections via EPHRIN-B/EPHB signaling
Jing Zhou, Yong Lin, Trung Huynh, Hirofumi Noguchi, Jeffrey O. Bush, Samuel J. Pleasure
MicroRNA-19b regulates proliferation and differentiation along the medial-lateral axis of the developing avian pallium
Suvimal Kumar Sindhu, Archita Mishra, Niveda Udaykumar, Jonaki Sen
Identification of ADAMTS19 as a novel retinal factor involved in ocular growth regulation
Swanand Koli, Cassandre Labelle-Dumais, Yin Zhao, Seyyedhassan Paylakhi, K Saidas Nair
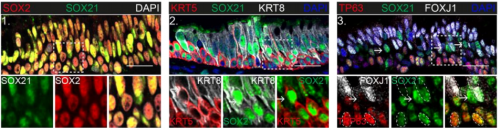
SOX21 modulates SOX2-initiated differentiation of epithelial cells in the extrapulmonary airways
Evelien Eenjes, Marjon Buscop-van Kempen, Anne Boerema-de Munck, Lisette de Kreij-de Bruin, J. Marco Schnater, Dick Tibboel, Jennifer J.P. Collins, Robbert J. Rottier
RUNX1 marks a luminal castration resistant lineage established at the onset of prostate development
Renaud Mevel, Ivana Steiner, Susan Mason, Laura Galbraith, Rahima Patel, Muhammad ZH Fadlullah, Imran Ahmad, Hing Y. Leung, Pedro Oliveira, Karen Blyth, Esther Baena, Georges Lacaud
Tbr2-expressing retinal ganglion cells are ipRGCs
Chai-An Mao, Ching-Kang Chen, Takae Kiyama, Nicole Weber, Christopher M. Whitaker, Ping Pan, Tudor C. Badea, Stephen C. Massey
Loss of coiled-coil protein Cep55 impairs abscission processes and results in p53-dependent apoptosis in developing cortex
Jessica N. Little, Katrina C. McNeely, Nadine Michel, Christopher J. Bott, Kaela S. Lettieri, Madison R. Hecht, Sara A. Martin, Noelle D. Dwyer
CROCCP2 acts as a human-specific modifier of cilia dynamics and mTOR signalling to promote expansion of cortical progenitors
Roxane Van Heurck, Marta Wojno, Ikuo K. Suzuki, Fausto D. Velez-Bravo, Jérôme Bonnefont, Emir Erkol, Dan Truc Nguyen, Adèle Herpoel, Angéline Bilheu, Catherine Ledent, Pierre Vanderhaeghen
Human SYNGAP1 Regulates the Development of Neuronal Activity by Controlling Dendritic and Synaptic Maturation
Nerea Llamosas, Vineet Arora, Ridhima Vij, Murat Kilinc, Lukasz Bijoch, Camilo Rojas, Adrian Reich, BanuPriya Sridharan, Erik Willems, David R. Piper, Louis Scampavia, Timothy P. Spicer, Courtney A. Miller, J. Lloyd Holder Jr, Gavin Rumbaugh
Comparison of human and mouse fetal intestinal tissues reveals differential maturation timelines
A.A. Lim, R.R. Nadkarni, B.C. Courteau, J.S. Draper
Targeted disruption of Pparγ1 promotes trophoblast endoreplication in the murine placenta
Takanari Nakano, Hidekazu Aochi, Masataka Hirasaki, Yasuhiro Takenaka, Koji Fujita, Hiroaki Soma, Hajime Kamezawa, Takahiro Koizumi, Akihiko Okuda, Takayuki Murakoshi, Akira Shimada, Ikuo Inoue
A temporal map of maternal immune activation-induced changes reveals a shift in neurodevelopmental timing and perturbed cortical development in mice
Cesar P. Canales, Myka L. Estes, Karol Cichewicz, Kartik Angara, John Paul Aboubechara, Scott Cameron, Kathryn Prendergast, Linda Su-Feher, Iva Zdilar, Ellie J. Kreun, Emma C. Connolly, Jin M. Seo, Jack B. Goon, Kathleen Farrelly, Tyler Stradleigh, Deborah van der List, Lori Haapanen, Judy Van de Water, Daniel Vogt, A. Kimberley McAllister, Alex S. Nord
Activation of mitochondria is an acute Akt-dependent response during osteogenic differentiation
C. Owen Smith, Roman A. Eliseev
Phox2a defines a developmental origin of the anterolateral system in mice and humans
R. Brian Roome, Farin B. Bourojeni, Bishakha Mona, Shima Rastegar-Pouyani, Raphael Blain, Annie Dumouchel, Charleen Salesse, W. Scott Thompson, Megan Brookbank, Yorick Gitton, Lino Tessarollo, Martyn Goulding, Jane E. Johnson, Marie Kmita, Alain Chédotal, Artur Kania
Harmonization of L1CAM Expression Facilitates Axon Outgrowth and Guidance of a Motor Neuron
Tessa Sherry, Hannah R. Nicholas, Roger Pocock
The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis
James Matthew Ragle, Abigail L. Aita, Kayleigh N. Morrison, Raquel Martinez-Mendez, Hannah N. Saeger, Guinevere A. Ashley, Londen C. Johnson, Katherine A. Schubert, Diane C. Shakes, Jordan D. Ward
Dynamic expression and localization of the LIN-2/7/10 protein scaffolding complex during C. elegans vulval development
Kimberley D. Gauthier, Christian E. Rocheleau
The conserved ASCL1/MASH-1 ortholog HLH-3 specifies sex-specific ventral cord motor neuron fate in C. elegans
Lillian M. Perez, Aixa Alfonso
PIG-1 MELK-dependent phosphorylation of nonmuscle myosin II promotes apoptosis through CES-1 Snail partitioning
Hai Wei, Eric J. Lambie, Daniel S. Osório, Ana X. Carvalho, Barbara Conradt
Raising the Connectome: the emergence of neuronal activity and behavior in C. elegans
Bradly J Alicea
Axin-mediated regulation of lifespan and muscle health in C. elegans involves AMPK-FOXO signaling
Avijit Mallick, Ayush Ranawade, Bhagwati P Gupta
Early C. elegans embryos modulate cell division timing to compensate for, and survive, the discordant conditions of a severe temperature gradient
Eric Terry, Bilge Birsoy, David Bothman, Marin Sigurdson, Pradeep M. Joshi, Carl Meinhart, Joel H. Rothman
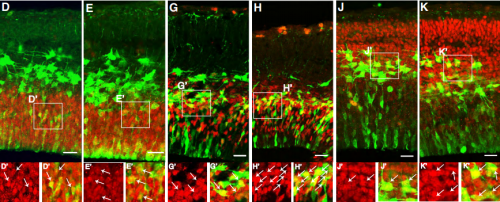
Neuralized regulates a travelling wave of Epithelium-to-Neural Stem Cell morphogenesis in Drosophila
Chloé Shard, Juan Luna-Escalante, François Schweisguth
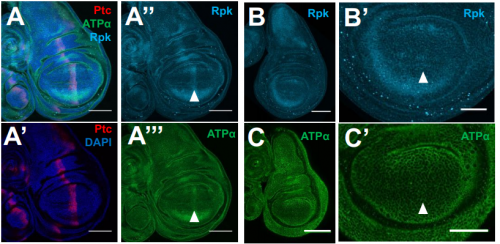
Membrane potential regulates Hedgehog signaling and compartment boundary maintenance in the Drosophila wing disc
Maya Emmons-Bell, Riku Yasutomi, Iswar K. Hariharan
Wnt ligands are not required for planar cell polarity in the Drosophila wing or notum
Ben Ewen-Campen, Typhaine Comyn, Eric Vogt, Norbert Perrimon
Feedback control of Wnt signaling based on histidine cluster co-aggregation between Naked/NKD and Axin
Melissa Gammons, Miha Renko, Joshua E. Flack, Juliusz Mieszczanek, Mariann Bienz
Multiple Wnts act synergistically to induce Chk1/Grapes expression and mediate G2 arrest in Drosophila tracheoblasts
Amrutha Kizhedathu, Rose Sebastian Kunnappallil, Archit V Bagul, Puja Verma, Arjun Guha
Balanced JAK/STAT signaling is critical to maintain the functional and structural integrity of the Drosophila respiratory epithelium
Xiao Niu, Christine Fink, Kimberley Kallsen, Viktoria Mincheva, Sören Franzenburg, Ruben Prange, Judith Bossen, Holger Heine, Thomas Roeder
Drosophila Hedgehog can act as a morphogen in the absence of regulated Ci processing
Jamie C. Little, Elisa Garcia-Garcia, Amanda Sul, Daniel Kalderon
Twist regulates Yorkie to guide lineage reprogramming of syncytial alary muscles
Marcel Rose, Jakob Bartle-Schultheis, Katrin Domsch, Christoph Schaub
fruitless tunes functional flexibility of courtship circuitry during development
Jie Chen, Sihui Jin, Jie Cao, Qionglin Peng, Yufeng Pan
| Morphogenesis & mechanics
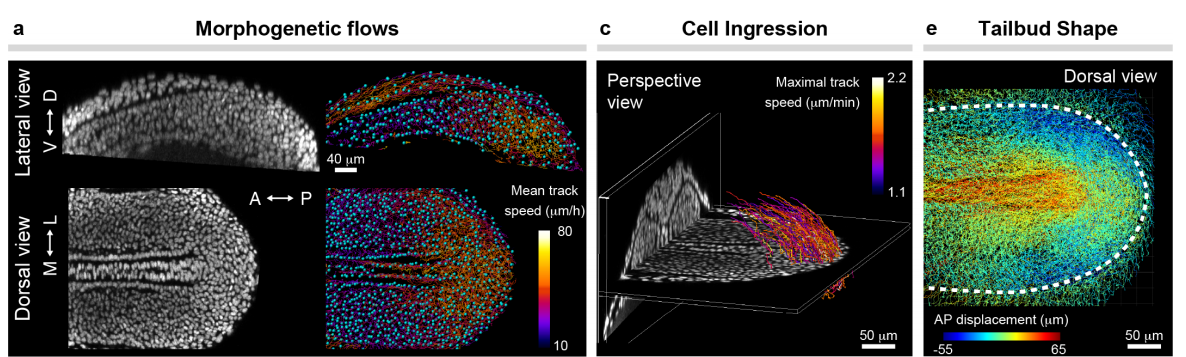
Mechanical control of tissue shape and morphogenetic flows during vertebrate body axis elongation
Samhita P. Banavar, Emmet K. Carn, Payam Rowghanian, Georgina Stooke-Vaughan, Sangwoo Kim, Otger Campàs
Embryonic Tissues as Active Foams
Sangwoo Kim, Marie Pochitaloff, Georgina-Stooke-Vaughan, Otger Campàs
Reconstituting Stratified Epithelial Branching Morphogenesis by Engineering Cell Adhesion
Shaohe Wang, Kazue Matsumoto, Kenneth M. Yamada
The Biomechanical Basis of Biased Epithelial Tube Elongation
Steve Runser, Lisa Conrad, Harold Gómez, Christine Lang, Mathilde Dumond, Aleksandra Sapala, Laura Kramps, Odysse Michos, Roman Vetter, Dagmar Iber
Nf2 fine-tunes proliferation and tissue alignment during closure of the optic fissure in the embryonic mouse eye
Wesley R. Sun, Sara Ramirez, Kelly E. Spiller, Yan Zhao, Sabine Fuhrmann
Integer topological defects organize stresses driving tissue morphogenesis
Pau Guillamat, Carles Blanch-Mercader, Karsten Kruse, Aurélien Roux
Hingepoints and neural folds reveal conserved features of primary neurulation in the zebrafish forebrain
Jonathan M Werner, Maraki Y Negesse, Dominique L Brooks, Allyson R Caldwell, Jafira M Johnson, Rachel Brewster
Sphingosine 1-phosphate activates the MAP3K1-JNK pathway to promote epithelial movement and morphogenesis
Jingjing Wang, Maureen Mongan, Jerold Chun, Ying Xia
Dentate gyrus development requires a cortical hem-derived astrocytic scaffold
Alessia Caramello, Christophe Galichet, Karine Rizzoti, Robin Lovell-Badge
Basal epidermis collective migration and local Sonic hedgehog signaling promote skeletal branching morphogenesis in zebrafish fins
Joshua A Braunstein, Amy E Robbins, Scott Stewart, Kryn Stankunas
Hapln1b organizes the ECM to modulate kit signaling and control developmental hematopoiesis in zebrafish
Christopher B. Mahony, Corentin Pasche, Vincent Braunersreuther, Savvas N. Savvides, Ariane de Agostini, Julien Y. Bertrand
Zebrafish follistatin-like 1b regulates cardiac contraction during early development
Xin-Xin I. Zeng, Karen Ocorr, Erik J. Ensberg, P. Duc si Dong
Requirement of Irf6 and Esrp1/2 in frontonasal and palatal epithelium to regulate craniofacial and palate morphogenesis in mouse and zebrafish
Shannon H. Carroll, Claudio Macias Trevino, Edward B-H Li, Kenta Kawasaki, Nora Alhazmi, Shawn Hallett, Justin Cotney, Russ P. Carstens, Eric C. Liao
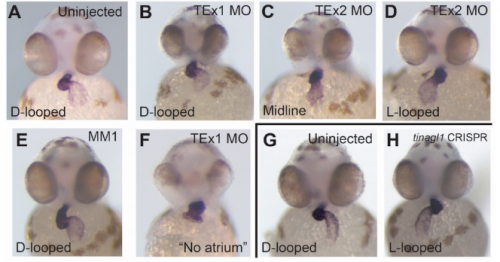
Acute knockdown of extracellular matrix protein Tinagl1 disrupts heart laterality and pronephric cilia in zebrafish embryonic development
Hannah Neiswender, Ellen K. LeMosy
Smooth muscle-specific MMP17 (MT4-MMP) defines the intestinal ECM niche
Mara Martín-Alonso, Håvard T. Lindholm, Sharif Iqbal, Pia Vornewald, Sigrid Hoel, Mirjam J. Damen, A.F.Maarten Altelaar, Pekka Katajisto, Alicia G. Arroyo, Menno J. Oudhoff
Notch Regulates Vascular Collagen IV Basement Membrane Through Modulation of Lysyl Hydroxylase 3 Trafficking
Stephen J. Gross, Amelia M. Webb, Alek D. Peterlin, Jessica R. Durrant, Rachel Judson, Erich J. Kushner
Syndecan-4-/- mice have smaller muscle fibers, increased Akt/mTOR/S6K1 and Notch/HES-1 pathways, and alterations in extracellular matrix components
Sissel Beate Rønning, Cathrine Rein Carlson, Jan Magnus Aronsen, Addolorata Pisconti, Vibeke Høst, Marianne Lunde, Kristian Hovde Liland, Ivar Sjaastad, Svein Olav Kolset, Geir Christensen, Mona Elisabeth Pedersen
Extracellular matrix protein composition dynamically changes during murine forelimb development
Kathryn R. Jacobson, Aya M. Saleh, Sarah N. Lipp, Alexander R. Ocken, Tamara L. Kinzer-Ursem, Sarah Calve
Prmt5 promotes vascular morphogenesis independently of its methyltransferase activity
Aurélie Quillien, Manon Boulet, Séverine Ethuin, Laurence Vandel
The Rab11 effectors Fip5 and Fip1 regulate zebrafish intestinal development
Cayla E. Jewett, Bruce H. Appel, Rytis Prekeris
Morphogenesis of the islets of Langerhans is guided by extra-endocrine Slit2/3 signals
Jennifer M. Gilbert, Melissa T. Adams, Nadav Sharon, Hariharan Jayaraaman, Barak Blum
Detecting new allies: Modifier screen identifies a genetic interaction between Imaginal disc growth factor 3 and a Rho-kinase substrate during dorsal appendage tube formation in Drosophila
Claudia Y. Espinoza, Celeste A. Berg
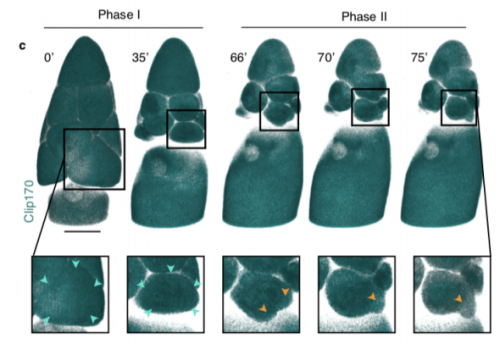
Dynamics of altruistic fluid transport in egg development
Alsous J Imran, N Romeo, J Jackson, FM Mason, J Dunkel, AC Martin
Post-mitotic myotubes repurpose the cytokinesis machinery to effect cellular guidance
Shuo Yang, Jennifer McAdow, Yingqiu Du, Jennifer Trigg, Paul H. Taghert, Aaron N. Johnson
FHOD-1 is the only formin in Caenorhabditis elegans that promotes striated muscle growth and Z-line organization in a cell autonomous manner
Sumana Sundaramurthy, SarahBeth Votra, Arianna Laszlo, Tim Davies, David Pruyne
Cell-extracellular matrix interactions in the fluidic phase direct the topology and polarity of self-organized epithelial structures
Mingxing Ouyang, Jiun-Yann Yu, Yenyu Chen, Linhong Deng, Chin-Lin Guo
A hydraulic instability drives the cell death decision in the nematode germline
N. T. Chartier, A. Mukherjee, J. Pfanzelter, S. Fürthauer, B. T. Larson, A.W. Fritsch, M. Kreysing, F. Jülicher, S. W. Grill
| Genes & genomes
Tissue-specific dynamic codon redefinition in Drosophila
Andrew M. Hudson, Gary Loughran, Nicholas L. Szabo, Norma M. Wills, John F. Atkins, Lynn Cooley
Deciphering the regulatory logic of a Drosophila enhancer through systematic sequence mutagenesis and quantitative image analysis
Yann Le Poul, Yaqun Xin, Liucong Ling, Bettina Mühling, Rita Jaenichen, David Hörl, David Bunk, Hartmann Harz, Heinrich Leonhardt, Yingfei Wang, Elena Osipova, Mariam Museridze, Deepak Dharmadhikari, Eamonn Murphy, Remo Rohs, Stephan Preibisch, Benjamin Prud’homme, Nicolas Gompel
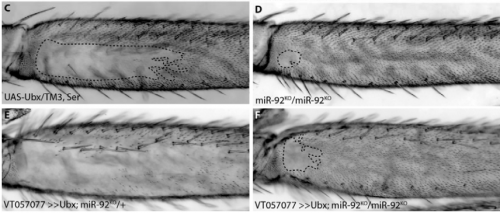
Characterisation of the role and regulation of Ultrabithorax in sculpting fine-scale leg morphology
Alexandra D. Buffry, Sebastian Kittelmann, Alistair P. McGregor
Collaboration between homologous recombination and non-homologous end joining in repair of meiotic double-strand breaks in Drosophila
Talia Hatkevich, Danny E. Miller, Carolyn A. Turcotte, Margaret C. Miller, Jeff Sekelsky
Modulation of the promoter activation rate dictates the transcriptional response to graded BMP signaling levels in the Drosophila embryo
Caroline Hoppe, Jonathan R. Bowles, Thomas G. Minchington, Catherine Sutcliffe, Priyanka Upadhyai, Magnus Rattray, Hilary L. Ashe
Excess histone H3 is a Chk1 inhibitor that controls embryonic cell cycle progression
Yuki Shindo, Amanda A. Amodeo
A mutation in the Drosophila melanogaster eve stripe 2 minimal enhancer is buffered by flanking sequences
Francheska Lopez-Rivera, Olivia K. Foster, Ben J. Vincent, Edward C. G. Pym, Meghan D. J. Bragdon, Javier Estrada, Angela H. DePace, Zeba Wunderlich
Germline inherited small RNAs clear untranslated maternal mRNAs in C. elegans embryos
Piergiuseppe Quarato, Meetali Singh, Eric Cornes, Blaise Li, Loan Bourdon, Florian Mueller, Celine Didier, Germano Cecere
Mutator foci are regulated by developmental stage, RNA, and the germline cell cycle in Caenorhabditis elegans
Celja J. Uebel, Dana Agbede, Dylan C. Wallis, Carolyn M. Phillips
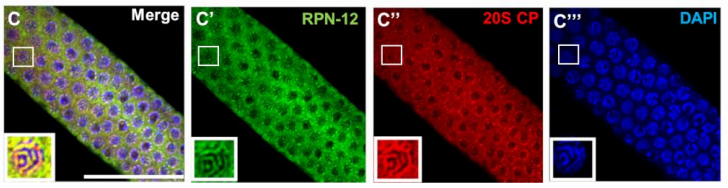
The C. elegans proteasome subunit RPN-12 is required for hermaphrodite germline sex determination and oocyte quality
Lourds M. Fernando, Jeandele Elliot, Anna K. Allen
Induction of RNA interference by C. elegans mitochondrial dysfunction via the DRH-1/RIG-I homologue RNA helicase and the EOL-1/RNA decapping enzyme
Kai Mao, Peter Breen, Gary Ruvkun
Two classes of active transcription sites and their roles in developmental regulation
Sarah Robinson-Thiewes, John McCloskey, Judith Kimble
Two microRNAs are sufficient for embryogenesis in C. elegans
Philipp J. Dexheimer, Jingkui Wang, Luisa Cochella
Otx2 and Oc1 directly regulate the transcriptional program of cone photoreceptor development
Nicolas Lonfat, Su Wang, ChangHee Lee, Jiho Choi, Peter J. Park, Constance Cepko
The RNA-binding protein Igf2bp3 is critical for embryonic and germline development in zebrafish
Yin Ho Vong, Lavanya Sivashanmugam, Andreas Zaucker, Alex Jones, Karuna Sampath
Chromatin remodeler Brahma safeguards canalization in cardiac mesoderm differentiation
Swetansu K. Hota, Andrew P. Blair, Kavitha S. Rao, Kevin So, Aaron M. Blotnick, Ravi V. Desai, Leor S. Weinberger, Irfan S. Kathiriya, Benoit G. Bruneau
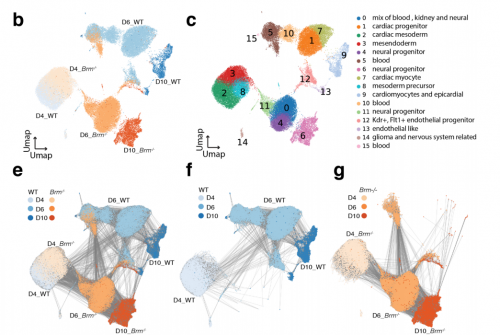
Chromatin and transcriptional response to loss of TBX1 in early differentiation of mouse cells
Andrea Cirino, Ilaria Aurigemma, Monica Franzese, Gabriella Lania, Dario Righelli, Rosa Ferrentino, Elizabeth Illingworth, Claudia Angelini, Antonio Baldini
NMDAR-mediated transcriptional control of gene expression in the specification of interneuron subtype identity
Vivek Mahadevan, Apratim Mitra, Yajun Zhang, Areg Peltekian, Ramesh Chittajallu, Caraoline Esnault, Dragan Maric, Christopher Rhodes, Kenneth A. Pelkey, Ryan Dale, Timothy J. Petros, Chris J. McBain
HMGXB4 Targets Sleeping Beauty Transposition to Vertebrate Germinal Stem Cells
Anantharam Devaraj, Manvendra Singh, Suneel Narayanavari, Guo Yong, Jiaxuan Wang, Jichang Wang, Mareike Becker, Oliver Walisko, Andrea Schorn, Zoltán Cseresznyés, Dawid Grzela, Tamás Raskó, Matthias Selbach, Zoltán Ivics, Zsuzsanna Izsvák
Fmr1 translationally activates stress-sensitive mRNAs encoding large proteins in oocytes and neurons
Ethan J. Greenblatt, Allan C. Spradling
An atlas of neural crest lineages along the posterior developing zebrafish at single-cell resolution
Aubrey G.A. Howard IV, Phillip A. Baker, Rodrigo Ibarra-García-Padilla, Joshua A. Moore, Lucia J. Rivas, Eileen W. Singleton, Jessa L. Westheimer, Julia A. Corteguera, James J. Tallman, Rosa A. Uribe
Scaling of gene transcriptional gradients with brain size across mouse development
Lau Hoi Yan Gladys, Alex Fornito, Ben D. Fulcher
The changing mouse embryo transcriptome at whole tissue and single-cell resolution
Peng He, Brian A. Williams, Diane Trout, Georgi K. Marinov, Henry Amrhein, Libera Berghella, Say-Tar Goh, Ingrid Plajzer-Frick, Veena Afzal, Len A. Pennacchio, Diane E. Dickel, Axel Visel, Bing Ren, Ross C. Hardison, Yu Zhang, Barbara J. Wold
Gene-environment interactions characterized by single embryo transcriptomics
Alfire Sidik, Groves B. Dixon, Hannah G. Kirby, Johann K. Eberhart
Single cell resolution regulatory landscape of the mouse kidney highlights cellular differentiation programs and renal disease targets
Zhen Miao, Michael S. Balzer, Ziyuan Ma, Hongbo Liu, Junnan Wu, Rojesh Shrestha, Tamas Aranyi, Amy Kwan, Ayano Kondo, Marco Pontoglio, Junhyong Kim, Mingyao Li, Klaus H. Kaestner, Katalin Susztak
Dynamic extrinsic pacing of the HOX clock in human axial progenitors controls motor neuron subtype specification
Vincent Mouilleau, Célia Vaslin, Simona Gribaudo, Rémi Robert, Nour Nicolas, Margot Jarrige, Angélique Terray, Léa Lesueur, Mackenzie W. Mathis, Gist Croft, Mathieu Daynac, Virginie Rouiller-Fabre, Hynek Wichterle, Vanessa Ribes, Cécile Martinat, Stéphane Nedelec
Differential abilities to engage inaccessible chromatin diversify vertebrate HOX binding patterns
Milica Bulajić, Divyanshi Srivastava, Jeremy S Dasen, Hynek Wichterle, Shaun Mahony, Esteban O Mazzoni
SPECIFIC ECTODERMAL ENHANCERS CONTROL THE EXPRESSION OF Hoxc GENES IN DEVELOPING MAMMALIAN INTEGUMENTS
Marc Fernandez-Guerrero, Nayuta Yakushiji-Kaminatsui, Lucille Lopez-Delisle, Sofía Zdral, Fabrice Darbellay, Rocío Perez-Gomez, Christopher Chase Bolt, Manuel A. Sanchez-Martin, Denis Duboule, Maria A. Ros
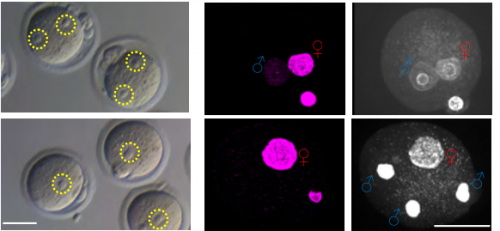
Histone H3.3 Hira chaperone complex contributes to zygote formation in mice and humans
Rowena Smith, Sue Pickering, Anna Kopakaki, K Joo Thong, Richard A Anderson, Chih-Jen Lin
The long noncoding RNA Meg3 regulates myoblast plasticity and muscle regeneration through epithelial-mesenchymal transition
Tiffany L. Dill, Alina Carroll, Jiachen Gao, Francisco J. Naya
Transcriptional heterogeneity of stemness phenotypes in the ovarian epithelium
LE. Carter, DP. Cook, CW. McCloskey, T. Dang, O. Collins, LF. Gamwell, HA. Dempster, BC. Vanderhyden
| Stem cells, regeneration & disease modelling
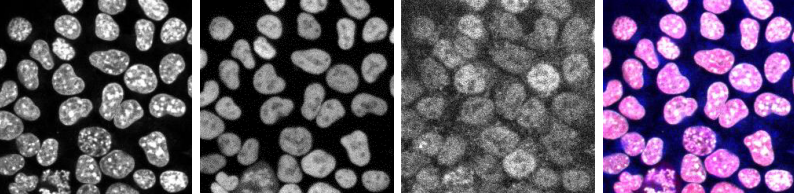
The combined action of Esrrb and Nr5a2 is essential for naïve pluripotency
Nicola Festuccia, Nick Owens, Almira Chervova, Agnès Dubois, Pablo Navarro
Intestinal progenitor P-bodies maintain stem cell identity by suppressing pro-differentiation factors
Kasun Buddika, Yi-Ting Huang, Alex Butrum-Griffith, Sam A. Norrell, Alex M. O’Connor, Viraj K. Patel, Samuel A. Rector, Mark Slovan, Mallory Sokolowski, Yasuko Kato, Akira Nakamura, Nicholas S. Sokol
Defining compartmentalized stem and progenitor populations with distinct cell division frequency in the ocular surface epithelium
Ryutaro Ishii, Hiromi Yanagisawa, Aiko Sada
Transcriptional networks are dynamically regulated during cell cycle progression in human Pluripotent Stem Cells
Anna Osnato, Ludovic Vallier
Epigenetic regulations follow cell cycle progression during differentiation of human pluripotent stem cells
Pedro Madrigal, Siim Pauklin, Kim Jee Goh, Rodrigo Grandy, Anna Osnato, Daniel Ortmann, Stephanie Brown, Ludovic Vallier
AP-2γ is Required for Maintenance of Pluripotent Mammary Stem Cells
Vivian W. Gu, Edward Cho, Dakota T. Thompson, Victoria C. Cassady, Nicholas Borcherding, Kelsey E. Koch, Vincent T. Wu, Allison W. Lorenzen, Mikhail V. Kulak, Trevor Williams, Weizhou Zhang, Ronald J. Weigel
Opposing Wnt and JAK-STAT signaling gradients define a stem cell domain by regulating spatially patterned cell division and differentiation at two borders
David Melamed, Daniel Kalderon
Gradual segregation of Adult Stem Cells and Niche cells during development from common precursors under the guidance of graded extracellular signals
Amy Reilein, Helen V. Kogan, Rachel Misner, Karen Sophia Park, Daniel Kalderon
Follicle Stem Cells (FSCs) in the Drosophila ovary; a critique of published studies defining the number, location and behavior of FSCs
Daniel Kalderon, David Melamed, Amy Reilein
Bi-compartmentalized stem cell organization of the corneal limbal niche
Olivia Farrelly, Yoko Suzuki-Horiuchi, Megan Brewster, Paola Kuri, Sixia Huang, Gabriella Rice, Jianming Xu, Tzvete Dentchev, Vivian Lee, Panteleimon Rompolas
ZFP982 confers mouse embryonic stem cell characteristics by regulating expression of Nanog, Zfp42 and Dppa3
Fariba Dehghanian, Patrick Piero Bovio, Zohreh Hojati, Tanja Vogel
Embryonic stem cells commit to differentiation by symmetric divisions following a variable lag period
Stanley E Strawbridge, Guy B Blanchard, Austin Smith, Hillel Kugler, Graziano Martello
Wnt- and Glutamate-receptors orchestrate stem cell dynamics and asymmetric cell division
Sergi Junyent, Joshua Reeves, James L. A. Szczerkowski, Clare L. Garcin, Tung-Jui Trieu, Matthew Wilson, Shukry J. Habib
Endogenous neural stem cells modulate microglia and protect from demyelination
Béatrice Brousse, Karine Magalon, Fabrice Daian, Pascale Durbec, Myriam Cayre
Mouse thy1-positive spermatogonia suppress the proliferation of spermatogonial stem cells by Extracellular vesicles in vitro
Yu Lin, Qian Fang, Yue He, Xiaowen Gong, Yinjuan Wang, Ajuan Liang, Guishuan Wang, Shengnan Gong, Ji Wu, Fei Sun
An ATM-MYBL2-CDC7 axis regulates replication initiation and prevents replication stress in pluripotent stem cells
Daniel Blakemore, Ruba Almaghrabi, Nuria Vilaplana, Elena Gonzalez, Miriam Moya, George Murphy, Grant Stewart, Agnieszka Gambus, Eva Petermann, Paloma García
CSF1R inhibition by a small molecule inhibitor affects hematopoiesis and the function of macrophages
Fengyang Lei, Naiwen Cui, Chengxin Zhou, James Chodosh, Demetrios G. Vavvas, Eleftherios I. Paschalis
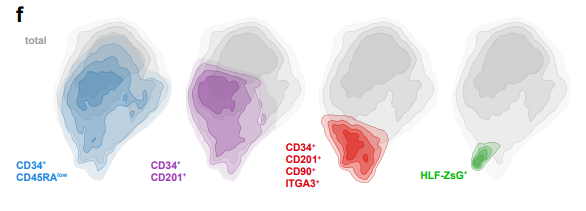
HLF Expression Defines the Human Haematopoietic Stem Cell State
Bernhard Lehnertz, Tara MacRae, Jalila Chagraoui, Elisa Tomellini, Sophie Corneau, Nadine Mayotte, Isabel Boivin, Guy Sauvageau
Modulation of Aplnr signaling is required during the development and maintenance of the hematopoietic system
Melany Jackson, Antonella Fidanza, A. Helen Taylor, Stanislav Rybtsov, Richard Axton, Maria Kydonaki, Stephen Meek, Tom Burdon, Alexander Medvinsky, Lesley M. Forrester
Spatial confinement and temporal dynamics of selectin ligands enable stable hematopoietic stem cell rolling
Bader Al Alwan, Karmen AbuZineh, Shuho Nozue, Aigerim Rakhmatulina, Mansour Aldehaiman, Asma S. Al-Amoodi, Maged F. Serag, Fajr A. Aleisa, Jasmeen S. Merzaban, Satoshi Habuchi
Selective linkage of mitochondrial enzymes to intracellular calcium stores differs between human induced pluripotent stem cells, neural stem cells and neurons
Huanlian Chen, Ankita Thakkar, Abigail Cross, Hui Xu, Aiqun Li, Daniel Pauli, Scott A. Noggle, Laken Kruger, Travis T. Denton, Gary E. Gibson
Orthotopic Transplantation and Engraftment of Human Induced Pluripotent Stem Cell-Derived Alveolar Progenitor Cells into Murine Lungs
Aaron I. Weiner, Rafael Fernandez, Gan Zhao, Gargi Palashikar, Maria Fernanda de Mello Costa, Stephanie Adams, Christopher J. Lengner, F. Brad Johnson, Andrew E. Vaughan
miR-203 imposes an intrinsic barrier during cellular reprogramming by targeting NFATC2
María Salazar-Roa, Sara Martínez-Martínez, Osvaldo Graña-Castro, Mónica Álvarez-Fernández, Marianna Trakala, Juan-Miguel Redondo, Marcos Malumbres
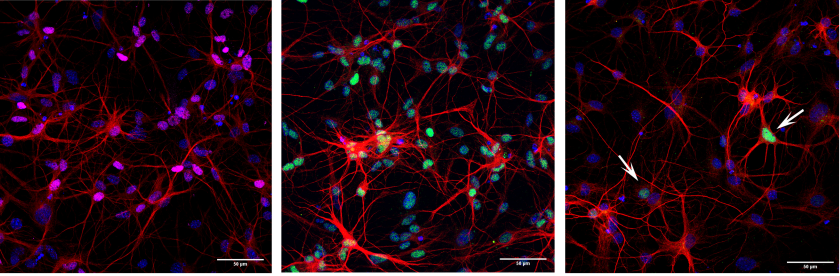
Cooperative action of miR-124 and ISX9 in instructing direct reprogramming of mouse astrocytes to induced-neurons in vitro and in vivo
Elsa Papadimitriou, Paraskevi N. Koutsoudaki, Timokratis Karamitros, Dimitra Karagkouni, Dafni Chroni-Tzartou, Maria Margariti, Christos Gkemisis, Evangelia Xingi, Irini Thanou, Socrates J. Tzartos, Artemis G. Hatzigeorgiou, Dimitra Thomaidou
Induction of Muscle Regenerative Multipotent Stem Cells from Human Adipocytes by PDGF-AB and 5-Azacytidine
Avani Yeola, Shruthi Subramanian, Rema A. Oliver, Christine A. Lucas, Julie A. I. Thoms, Feng Yan, Jake Olivier, Diego Chacon, Melinda L. Tursky, Tzongtyng Hung, Carl Power, Philip Hardy, David D. Ma, Joshua McCarroll, Maria Kavallaris, Luke B. Hesson, Dominik Beck, David J. Curtis, Jason W.H. Wong, Edna C. Hardeman, William R. Walsh, Ralph Mobbs, Vashe Chandrakanthan, John E. Pimanda
Mms19 promotes spindle microtubule assembly in neural stem cells through two distinct pathways
Rohan Chippalkatti, Boris Egger, Beat Suter
Functional Characterization of the Lin28/let-7 Circuit during Forelimb Regeneration in Ambystoma mexicanum and its Influence on Metabolic Reprogramming
Hugo Varela-Rodríguez, Diana G. Abella-Quintana, Luis Varela-Rodríguez, David Gomez-Zepeda, Annie Espinal-Centeno, Juan Caballero-Pérez, José Juan Ordaz-Ortiz, Alfredo Cruz-Ramírez
Secreted inhibitors drive the loss of regeneration competence in Xenopus limbs
C. Aztekin, T. W. Hiscock, J. B. Gurdon, J. Jullien, J. C. Marioni, B. D. Simons
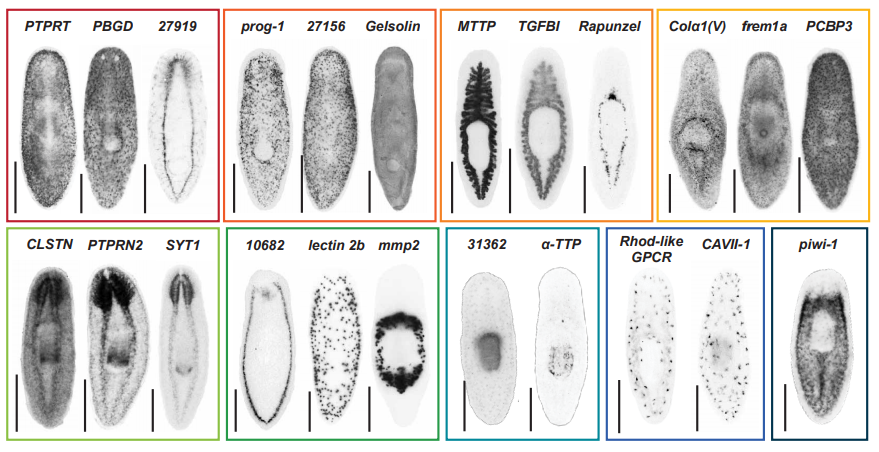
Identification of rare transient somatic cell states induced by injury and required for whole-body regeneration
Blair W. Benham-Pyle, Carolyn E. Brewster, Aubrey M. Kent, Frederick G. Mann Jr., Shiyuan Chen, Allison R. Scott, Andrew C. Box, Alejandro Sánchez Alvarado
Salamander-like tail regeneration in the West African lungfish
Kellen Matos Verissimo, Louise Neiva Perez, Aline Cutrim Dragalzew, Gayani Senevirathne, Sylvain Darnet, Wainna Renata Barroso Mendes, Ciro Ariel dos Santos Neves, Erika Monteiro dos Santos, Cassia Nazare de Sousa Moraes, Ahmed Elewa, Neil Shubin, Nadia Belinda Froebisch, Josane de Freitas Sousa, Igor Schneider
Mechanosensory neuron regeneration in adult Drosophila
Ismael Fernández-Hernández, Evan B. Marsh, Michael A. Bonaguidi
Regenerative neurogenic response from glia requires insulin driven neuron-glia communication
Neale Harrison, Elizabeth Connolly, Alicia Gascón Gubieda, Zidan Yang, Benjamin Altenhein, Maria Losada-Perez, Marta Moreira, Alicia Hidalgo
Diverse Epithelial Cell Populations Contribute to Regeneration of Secretory Units in Injured Salivary Glands
Ninche Ninche, MinGyu Kwak, Soosan Ghazizadeh
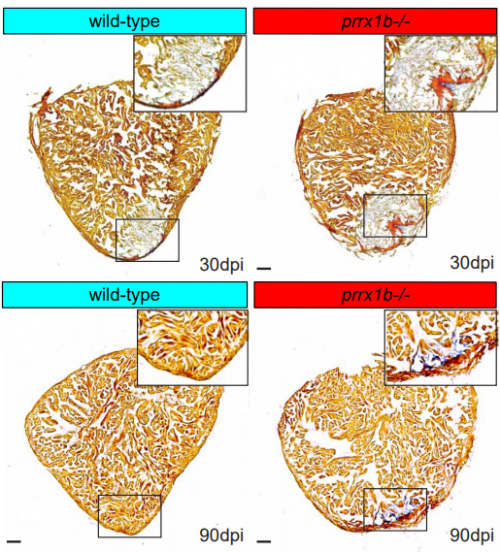
Prrx1b directs pro-regenerative fibroblasts during zebrafish heart regeneration
Dennis E.M. de Bakker, Esther Dronkers, Mara Bouwman, Aryan Vink, Marie-José Goumans, Anke M. Smits, Jeroen Bakkers
LIN28B controls the regenerative capacity of neonatal murine auditory supporting cells through activation of mTOR signaling
Xiaojun Li, Angelika Doetzlhofer
Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds
Quan M. Phan, Gracelyn Fine, Lucia Salz, Gerardo G. Herrera, Ben Wildman, Iwona M. Driskell, Ryan R. Driskell
Age-related degeneration leads to gliosis but not regeneration in the zebrafish retina
Raquel R Martins, Mazen Zamzam, Mariya Moosajee, Ryan B Thummel, Catarina M Henriques, Ryan B MacDonald
Reconstitution of Alveolar Regeneration via novel DATPs by Inflammatory Niches
Jinwook Choi, Jong-Eun Park, Georgia Tsagkogeorga, Motoko Yanagita, Bon-Kyoung Koo, Namshik Han, Joo-Hyeon Lee
b3galt6 knock-out zebrafish recapitulate β3GalT6-deficiency disorders in human and reveal a trisaccharide proteoglycan linkage region
Sarah Delbaere, Adelbert De Clercq, Shuji Mizumoto, Fredrik Noborn, Jan Willem Bek, Lien Alluyn, Charlotte Gistelinck, Delfien Syx, Phil L. Salmon, Paul J. Coucke, Göran Larson, Shuhei Yamada, Andy Willaert, Fransiska Malfait
Ciliopathic micrognathia is caused by aberrant skeletal differentiation and remodeling
Christian Louis Bonatto-Paese, Evan C Brooks, Megan Aarnio-Peterson, Samantha A Brugmann
Cdon mutation and fetal alcohol converge on Nodal signaling in a mouse model of holoprosencephaly
Mingi Hong, Annabel Christ, Anna Christa, Thomas E. Willnow, Robert S. Krauss
ALX1-related Frontonasal Dysplasia Results From Defective Neural Crest Cell Development and Migration
Jonathan Pini, Janina Kueper, Yiyuan David Hu, Kenta Kawasaki, Pan Yeung, Casey Tsimbal, Baul Yoon, Nikkola Carmichael, Richard L. Maas, Justin Cotney, Yevgenya Grinblat, Eric C. Liao
Patient-specific functional genomics and disease modeling suggest a role for LRP2 in hypoplastic left heart syndrome
Jeanne L. Theis, Georg Vogler, Maria A. Missinato, Xing Li, Almudena Martinez-Fernandez, Tanja Nielsen, Stanley M. Walls, Anais Kervadec, Xin-Xin I Zeng, James N. Kezos, Katja Birker, Jared M. Evans, Megan M. O’Byrne, Zachary C. Fogarty, André Terzic, Paul Grossfeld, Karen Ocorr, Timothy J. Nelson, Timothy M. Olson, Alexandre R. Colas, Rolf Bodmer
Loss of O-GlcNAcylation on MeCP2 Thr 203 Leads to Neurodevelopmental Disorders
Juanxian Cheng, Zhe Zhao, Liping Chen, Ruijing Du, Yan Wu, Qian Zhu, Ming Fan, Xiaotao Duan, Haitao Wu
Haploinsufficiency of the psychiatric risk gene Cyfip1 causes abnormal postnatal hippocampal neurogenesis through microglial and Arp2/3 mediated actin dependent mechanisms
Niels Haan, Laura J Westacott, Jenny Carter, Michael J Owen, William P Gray, Jeremy Hall, Lawrence S Wilkinson
Mosaic expression of X-linked PCDH19 Protein by in Utero Electroporation in Rats Replicates Human Cortical and Hippocampal Developmental Abnormalities, Associated Core Behaviors Related to Autism, and Cognitive Impairment
Andrzej W Cwetsch, Roberto Narducci, Maria Bolla, Bruno Pinto, Laura Perlini, Silvia Bassani, Maria Passafaro, Laura Cancedda
Cortical Organoids Model Early Brain Development Disrupted by 16p11.2 Copy Number Variants in Autism
Jorge Urresti, Pan Zhang, Patricia Moran-Losada, Nam-Kyung Yu, Priscilla D. Negraes, Cleber A. Trujillo, Danny Antaki, Megha Amar, Kevin Chau, Akula Bala Pramod, Jolene Diedrich, Leon Tejwani, Sarah Romero, Jonathan Sebat, John R. Yates III, Alysson R. Muotri, Lilia M. Iakoucheva
| Plant development
SCARECROW gene function is required for photosynthetic development in maize
Thomas E Hughes, Jane A Langdale
A single cell Arabidopsis root atlas reveals developmental trajectories in wild type and cell identity mutants
Rachel Shahan, Che-Wei Hsu, Trevor M. Nolan, Benjamin J. Cole, Isaiah W. Taylor, Anna Hendrika Cornelia Vlot, Philip N. Benfey, Uwe Ohler
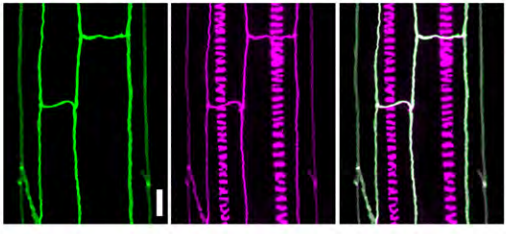
High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification
Nelson Rojas-Murcia, Kian Hématy, Yuree Lee, Aurélia Emonet, Robertas Ursache, Satoshi Fujita, Damien De Bellis, Niko Geldner

Tissue-autonomous phenylpropanoid production is essential for establishment of root barriers
Tonni Grube Andersen, David Molina, Joachim Kilian, Rochus Franke, Laura Ragni, Niko Geldner
The Arabidopsis R-SNARE VAMP714 is essential for polarization of PIN proteins in the establishment and maintenance of auxin gradients
Xiaoyan Gu, Kumari Fonseka, Stuart A. Casson, Andrei Smertenko, Guangqin Guo, Jennifer F. Topping, Patrick J. Hussey, Keith Lindsey
The Arabidopsis NRT1/PTR FAMILY Protein NPF7.3/NRT1.5 is an Indole-3-butyric Acid Transporter Involved in Root Gravitropism
Shunsuke Watanabe, Naoki Takahashi, Yuri Kanno, Hiromi Suzuki, Yuki Aoi, Noriko Takeda-Kamiya, Kiminori Toyooka, Hiroyuki Kasahara, Ken-Ichiro Hayashi, Masaaki Umeda, Mitsunori Seo
Flavonols modulate lateral root emergence by scavenging reactive oxygen species in Arabidopsis thaliana
Jordan M. Chapman, Gloria K. Muday
Expansin-controlled cell wall stiffness regulates root growth in Arabidopsis
Marketa Samalova, Kareem Elsayad, Alesia Melnikava, Alexis Peaucelle, Evelina Gahurova, Jaromir Gumulec, Ioannis Spyroglou, Elena V. Zemlyanskaya, Elena V. Ubogoeva, Jan Hejatko
GDSL-domain containing proteins mediate suberin biosynthesis and degradation, enabling developmental plasticity of the endodermis during lateral root emergence
Robertas Ursache, Cristovao De Jesus Vieira-Teixeira, Valérie Dénervaud Tendon, Kay Gully, Damien De Bellis, Emanuel Schmid-Siegert, Tonni Grube Andersen, Vinay Shekhar, Sandra Calderon, Sylvain Pradervand, Christiane Nawrath, Niko Geldner, Joop E.M. Vermeer
Phosphoproteomics after nitrate treatments reveal an important role for PIN2 phosphorylation in control of root system architecture
Andrea Vega, Isabel Fredes, José O’Brien, Zhouxin Shen, Krisztina Ötvös, Eva Benkova, Steven P. Briggs, Rodrigo A. Gutiérrez
The role of trehalose 6-phosphate in shoot branching – local and non-local effects on axillary bud outgrowth in arabidopsis rosettes
Franziska Fichtner, Francois F. Barbier, Maria G. Annunziata, Regina Feil, Justyna J. Olas, Bernd Mueller-Roeber, Mark Stitt, Christine A. Beveridge, John E. Lunn
Arabidopsis mTERF9 protein promotes chloroplast ribosomal assembly and translation by establishing ribonucleoprotein interactions in vivo
Louis-Valentin Méteignier, Rabea Ghandour, Aude Zimmerman, Lauriane Kuhn, Jörg Meurer, Reimo Zoschke, Kamel Hammani
A shortcut in forward genetics: concurrent discovery of mutant phenotype and causal mutation in Arabidopsis M2 families via MAD-mapping
Danalyn R Holmes, Robert Morbitzer, Markus Wunderlich, Hequan Sun, Farid El Kasmi, Korbinian Schneeberger, Thomas Lahaye
Multiple epigenetic layers accompany the spatial distribution of ribosomal genes in Arabidopsis
Konstantin O. Kutashev, Michal Franek, Klev Diamanti, Jan Komorowski, Marie Olšinová, Martina Dvořáčková
Exogenous Nitro-Oleic Acid inhibits primary root growth by reducing the mitosis in the meristem in Arabidopsis thaliana
Luciano M. Di Fino, Ignacio Cerrudo, Sonia R. Salvatore, Francisco J. Schopfer, Carlos García-Mata, Ana M. Laxalt
Requirement for proper mitochondrial RNA processing in the restrictive control of cell proliferation during early lateral root morphogenesis
Kurataka Otsuka, Akihito Mamiya, Mineko Konishi, Mamoru Nozaki, Atsuko Kinoshita, Hiroaki Tamaki, Masaki Arita, Masato Saito, Kayoko Yamamoto, Takushi Hachiya, Ko Noguchi, Takashi Ueda, Yusuke Yagi, Takehito Kobayashi, Takahiro Nakamura, Yasushi Sato, Takashi Hirayama, Munetaka Sugiyama
The Receptor Kinase BRI1 promotes cell proliferation in Arabidopsis by phosphorylation- mediated inhibition of the growth repressing peptidase DA1
Hui Dong, Caroline Smith, Rachel Prior, Ross Carter, Jack Dumenil, Gerhard Saalbach, Neil McKenzie, Michael Bevan
ASYMMETRIC EXPRESSION OF ARGONAUTES IN ARABIDOPSIS REPRODUCTIVE TISSUES
PE Jullien, DMV Bonnet, N Pumplin, JA Schröder, O Voinnet
Modulation of root growth by nutrient-defined fine-tuning of polar auxin transport
Krisztina Otvos, Marco Marconi, Andrea Vega, Jose O’Brien, Alexander Johnson, Rashed Abualia, Livio Antonielli, Juan Carlos Montesinos, Yuzhou Zhang, Shu-Tang Tan, Candela Cuesta, Christina Artner, Eleonore Bouguyon, Alain Gojon, Jiri Friml, Rodrigo A Gutiérrez, Krzysztof Wabnik, Eva Benková
Xyloglucan remodelling defines differential tissue expansion in plants
Silvia Melina Velasquez, Xiaoyuan Guo, Marçal Gallemi, Bibek Aryal, Peter Venhuizen, Elke Barbez, Kai Dünser, Martin Darino, Aleč Pěnčik, Ondřej Novák, Maria Kalyna, Grégory Mouille, Eva Benkova, Rishikesh Bhalerao, Jozef Mravec, Jürgen Kleine-Vehn
A novel pathway controlling cambium initiation and – activity via cytokinin biosynthesis in Arabidopsis
Arezoo Rahimi, Omid Karami, Angga Dwituti Lestari, Dongbo Shi, Thomas Greb, Remko Offringa
Mutations in Tomato ACC Synthase2 Uncover Its Role in Development beside Fruit Ripening
Kapil Sharma, Soni Gupta, Supriya Sarma, Meenakshi Rai, Yellamaraju Sreelakshmi, Rameshwar Sharma
The framework of lncRNAs and genes at early pollen developmental stage in a PTGMS wheat line
Jian-fang Bai, Zi-han Liu, Yu-kun Wang, Hao-yu Guo, Li-Ping Guo, Zhao-guo Tan, Shao-hua Yuan, Yan-mei Li, Ting-ting Li, Wen-jing Duan, Jie-ru Yue, Feng-ting Zhang, Chang-ping Zhao, Li-ping Zhang
Cytokinin-promoted secondary growth and nutrient storage in the perennial stem zone of Arabis alpina
Anna Sergeeva, Hongjiu Liu, Hans-Jörg Mai, Tabea Mettler-Altmann, Christiane Kiefer, George Coupland, Petra Bauer
Genome-Wide High Resolution Expression Map and Functions of Key Cell Fate Determinants Reveal the Dynamics of Crown Root Development in Rice
Tushar Garg, Zeenu Singh, Anuj K. Dwivedi, Vijina Varapparambathu, Raj Suryan Singh, Manoj Yadav, Divya Chandran, Kalika Prasad, Mukesh Jain, Shri Ram Yadav
Pre-meiotic, 24-nt reproductive phasiRNAs are abundant in anthers of wheat and barley but not rice and maize
Sébastien Bélanger, Suresh Pokhrel, Kirk Czymmek, Blake C. Meyers
The regulatory landscape of early maize inflorescence development
Rajiv K. Parvathaneni, Edoardo Bertolini, Md Shamimuzzaman, Daniel Vera, Pei-Yau Lung, Brian R. Rice, Jinfeng Zhang, Patrick J. Brown, Alexander E. Lipka, Hank W. Bass, Andrea L. Eveland
Organogenesis and Vasculature of Anaxagorea and its Implications for the Integrated Axial-Foliar Origin of Angiosperm Carpel
Ya Li, Wei Du, Shuai Wang, Xiao-Fan Wang
VipariNama: RNA vectors to rapidly reprogram plant morphology and metabolism
Arjun Khakhar, Cecily Wang, Ryan Swanson, Sydney Stokke, Furva Rizvi, Surbhi Sarup, John Hobbs, Daniel F. Voytas
CRISPR-finder: A high throughput and cost effective method for identifying successfully edited A. thaliana individuals
Efthymia Symeonidi, Julian Regalado, Rebecca Schwab, Detlef Weigel
Evo-devo & evo
Unravelling the genetic basis for the rapid diversification of male genitalia between Drosophila species
Joanna F. D. Hagen, Cláudia C. Mendes, Shamma R. Booth, Javier Figueras Jimenez, Kentaro M. Tanaka, Franziska A. Franke, Luis. Baudouin-Gonzalez, Amber M. Ridgway, Saad Arif, Maria D. S. Nunes, Alistair P. McGregor

The evolution of Sox gene repertoires and regulation of segmentation in arachnids
Luis Baudouin-Gonzalez, Anna Schoenauer, Amber Harper, Grace Blakeley, Michael Seiter, Saad Arif, Lauren Sumner-Rooney, Steven Russell, Prashant P. Sharma, Alistair P. McGregor
Distinct genetic architectures underlie divergent thorax, leg, and wing pigmentation between Drosophila elegans and D. gunungcola
Jonathan H Massey, Jun Li, David Stern, Patricia Wittkopp
Variation in a pleiotropic hub gene drives morphological evolution: Insights from interspecific differences in head shape and eye size in Drosophila
Elisa Buchberger, Anıl Bilen, Sanem Ayaz, David Salamanca, Cristina Matas de las Heras, Armin Niksic, Isabel Almudi, Montserrat Torres-Oliva, Fernando Casares, Nico Posnien
The gene cortex controls scale colour identity in Heliconius
Luca Livraghi, Joseph J. Hanly, Ling Sheng Loh, Anna Ren, Ian A. Warren, Carolina Concha, Charlotte Wright, Jonah M. Walker, Jessica Foley, Henry Arenas-Castro, Lucas Rene Brenes, Arnaud Martin, W. Owen McMillan, Chris D. Jiggins
Deep origins for the tectal visual processing centers in chordates
Cezar Borba, Shea Schwennicke, Matthew J. Kourakis, William C. Smith

Evolution of the potassium channel gene Kcnj13 underlies colour pattern diversification in Danio fish
Marco Podobnik, Hans Georg Frohnhöfer, Christopher M. Dooley, Anastasia Eskova, Christiane Nüsslein-Volhard, Uwe Irion
Crosstalk between Nitric Oxide and Retinoic Acid pathways is essential for amphioxus pharynx development
F Caccavale, G Annona, L Subirana, H Escriva, S Bertrand, S D’Aniello
The comprehensive ontology of the anatomy and development of the solitary ascidian Ciona: the swimming larva and its metamorphosis
Kohji Hotta, Delphine Dauga, Lucia Manni
Establishment of an in vitro culture system to study the developmental biology (growth, mating and nodule formation) of Onchocerca volvulus with implications for anti-onchocerca drug discovery and screening
Narcisse Victor T. Gandjui, Abdel Jelil Njouendou, Eric Njih Gemeg, Fanny Fri Fombad, Manuel Ritter, Chi Anizette Kien, Valerine C. Chunda, Jerome Fru, Mathias E. Esum, Marc P. Hübner, Peter A. Enyong, Achim Hoerauf, Samuel Wanji
Opposing directions of stage-specific body length change in a close relative of C. elegans
Eric W. Hammerschmith, Gavin C. Woodruff, Patrick C. Phillips
A single nucleotide change underlies the genetic assimilation of a plastic trait
Paul Vigne, Clotilde Gimond, Celine Ferrari, Anne Vielle, Johan Hallin, Ania Pino-Querido, Sonia El Mouridi, Christian Frøkjær-Jensen, Thomas Boulin, Henrique Teotonio, Christian Braendle
A flagellate-to-amoeboid switch in the closest living relatives of animals
Thibaut Brunet, Marvin Albert, William Roman, Danielle C. Spitzer, Nicole King
Interplay of mesoscale physics and agent-like behaviors in the parallel evolution of aggregative multicellularity
Juan A. Arias Del Angel, Vidyanand Nanjundiah, Mariana Benítez, Stuart A. Newman
Evolution of colonial life history in styelids tunicates involves changes in complexity patterns
Stefania Gutierrez
Functional characterization of a “plant-like” HYL1 homolog in the cnidarian Nematostella vectensis indicates a conserved involvement in microRNA biogenesis
Abhinandan Mani Tripathi, Arie Fridrich, Magda Lewandowska, Yehu Moran
Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri
Nathan J Kenny, Warren R. Francis, Ramón E. Rivera-Vicéns, Ksenia Juravel, Alex de Mendoza, Cristina Díez-Vives, Ryan Lister, Luis Bezares-Calderon, Lauren Grombacher, Maša Roller, Lael D. Barlow, Sara Camilli, Joseph F. Ryan, Gert Wörheide, April L Hill, Ana Riesgo, Sally P. Leys
Evolutionary transcriptomics implicates HAND2 in the origins of implantation and regulation of gestation length
Mirna Marinić, Katelyn Mika, Sravanthi Chigurupati, Vincent J. Lynch
Mutation bias shapes gene evolution in Arabidopsis thaliana
J. Grey Monroe, Thanvi Srikant, Pablo Carbonell-Bejerano, Moises Exposito-Alonso, Mao-Lun Weng, Matthew T. Rutter, Charles B. Fenster, Detlef Weigel
Meiosis reveals the early steps in the evolution of a neo-XY sex chromosome pair in the African pygmy mouse Mus minutoides
Ana Gil-Fernández, Paul Saunders, Marta Martín-Ruiz, Marta Ribagorda, Pablo López-Jiménez, Daniel L. Jeffries, María Teresa Parra, Aberto Viera, Julio S. Rufas, Nicolas Perrin, Frederic Veyrunes, Jesus Page
ZZ Top: faster and more adaptive Z chromosome evolution in two Lepidoptera
Andrew J. Mongue, Megan E. Hansen, James R. Walters
Haplotype tagging reveals parallel formation of hybrid races in two butterfly species
Joana I. Meier, Patricio A. Salazar, Marek Kučka, Robert William Davies, Andreea Dréau, Ismael Aldás, Olivia Box Power, Nicola J. Nadeau, Jon R. Bridle, Campbell Rolian, Nicholas H. Barton, W. Owen McMillan, Chris D. Jiggins, Yingguang Frank Chan
Phenotypes to remember: Evolutionary developmental memory capacity and robustness
András Szilágyi, Péter Szabó, Mauro Santos, Eörs Szathmáry
Evolution of sperm morphology in Daphnia
Duneau David, Markus Möst, Dieter Ebert
Cerebellar nuclei evolved by repeatedly duplicating a conserved cell type set
Justus M Kebschull, Noam Ringach, Ethan B Richman, Drew Friedmann, Sai Saroja Kolluru, Robert C Jones, William E Allen, Ying Wang, Huaijun Zhou, Seung Woo Cho, Howard Y Chang, Karl Deisseroth, Stephen R Quake, Liqun Luo
Cell biology
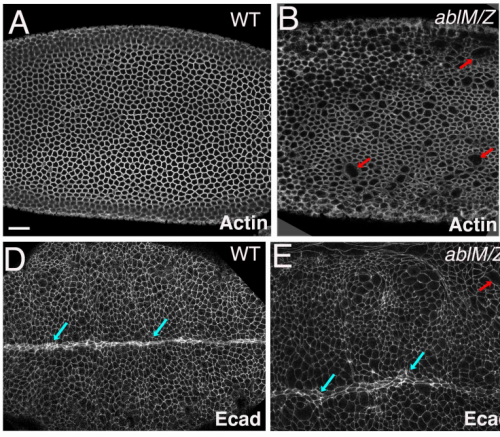
Abelson kinases intrinsically disordered linker plays important roles in protein function and protein stability
Edward M. Rogers, S. Colby Allred, Mark Peifer
Identification of the critical replication targets of CDK reveals direct regulation of replication initiation factors by the embryo polarity machinery in C. elegans
Vincent Gaggioli, Manuela R. Kieninger, Anna Klucnika, Richard Butler, Philip Zegerman

Co-movement of astral microtubules, organelles and F-actin suggests aster positioning by surface forces in frog eggs
James Pelletier, Christine Field, Sebastian Fürthauer, Matthew Sonnett, Timothy Mitchison
Organization of DNA replication origin firing in Xenopus egg extracts : the role of intra-S checkpoint
Diletta Ciardo, Olivier Haccard, Hemalatha Narassimprakash, Jean-Michel Arbona, Olivier Hyrien, Benjamin Audit, Kathrin Marheineke, Arach Goldar
Initial spindle formation at the oocyte center protects against incorrect kinetochore-microtubule attachment and aneuploidy in mice
Jessica N. Kincade, Ahmed Z. Balboula
Cytoplasmic Microtubule Organizing Centers Regulate Meiotic Spindle Positioning in Mouse Oocyte
Daniela Londono Vasquez, Katherine Rodriguez-Lukey, Susanta K. Behura, Ahmed Z. Balboula
PP2A:B56 Regulates Meiotic Chromosome Segregation in C. elegans Oocytes
Laura Bel Borja, Flavie Soubigou, Samuel J.P. Taylor, Conchita Fraguas Bringas, Jacqueline Budrewicz, Pablo Lara-Gonzalez, Christopher G. Sorensen-Turpin, Joshua N. Bembenek, Dhanya K. Cheerambathur, Federico Pelisch
Live imaging of chromatin distribution in muscle nuclei reveals novel principles of nuclear architecture and chromatin compartmentalization
Daria Amiad Pavlov, Dana Lorber, Gaurav Bajpai, Samuel Safran, Talila Volk
In vivo characterisation of endogenous cardiovascular extracellular vesicles in larval and adult zebrafish
Aaron Scott, Lorena Sueiro Ballesteros, Marston Bradshaw, Ann Power, James Lorriman, John Love, Danielle Paul, Andrew Herman, Costanza Emanueli, Rebecca J. Richardson
The SPIRE1 actin nucleator coordinates actin/myosin functions in the regulation of mitochondrial motility
Felix Straub, Tobias Welz, Hannah Alberico, Rafael Oliveira Brandão, Anna Huber, Annette Samol-Wolf, Cord Brakebusch, Dori Woods, Martin Kollmar, Javier Martin-Gonzalez, Eugen Kerkhoff
Spindle scaling is governed by cell boundary regulation of microtubule nucleation
Elisa Maria Rieckhoff, Frederic Berndt, Stefan Golfier, Franziska Decker, Maria Elsner, Keisuke Ishihara, Jan Brugués
Chromosome-directed oocyte spindle assembly depends HP1 and the Chromosomal Passenger Complex
Lin-Ing Wang, Tyler DeFosse, Rachel A. Battaglia, Victoria F. Wagner, Kim S. McKim
Distinct Roles of Tumor-Associated Mutations in Collective Cell Migration
Rachel M. Lee, Michele I. Vitolo, Wolfgang Losert, Stuart S. Martin
Mutational inactivation of Apc in the intestinal epithelia compromises cellular organisation
Helena Rannikmae, Samantha Peel, Simon Barry, Inderpreet Sur, Jussi Taipale, Takao Senda, Marc de la Roche
Keratins couple with the nuclear lamina and regulate proliferation in colonic epithelial cells
Carl-Gustaf A. Stenvall, Joel H. Nyström, Ciarán Butler-Hallissey, Stephen A. Adam, Roland Foisner, Karen M. Ridge, Robert D. Goldman, Diana M. Toivola
Modelling
Four different mechanisms for switching cell polarity
Filipe Tostevin, Manon Wigbers, Lotte Søgaard-Andersen, Ulrich Gerland
A quantitative principle to understand 3D cellular connectivity in epithelial tubes
Pedro Gomez-Galvez, Pablo Vicente-Munuera, Samira Anbari, Antonio Tagua, Carmen Gordillo, Ana Maria Palacios, Antonio Velasco, Carlos Capitan-Agudo, Clara Grima, Valentina Annese, Rafael Robles, Alberto Marquez, Javier Buceta, Luis M. Escudero
Computational modelling unveils how epiblast remodelling and positioning rely on trophectoderm morphogenesis during mouse implantation
Joel Dokmegang, Moi Hoon Yap, Liangxiu Han, Matteo Cavaliere, René Doursat
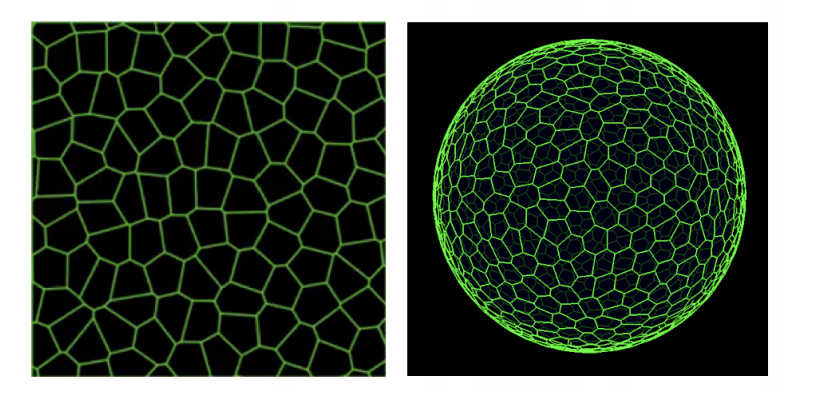
Interplay of curvature and rigidity in shape-based models of confluent tissue
Daniel M. Sussman
Diffusion vs. direct transport in the precision of morphogen readout
Sean Fancher, Andrew Mugler
A Biophysical Model for Plant Cell Plate Development
Muhammad Zaki Jawaid, Rosalie Sinclair, Daniel Cox, Georgia Drakakaki
Dynamics of a cell motility model near the sharp interface limit
Nicolas Bolle, Matthew S. Mizuhara
A mathematical model of cell fate selection on a dynamic tissue
Domenic P.J. Germano, James M. Osborne
Stick-Slip model for actin-driven cell protrusions, cell polarisation and crawling
Pierre Sens
Tools & resources
Stable integration of an optimized inducible promoter system enables spatiotemporal control of gene expression throughout avian development
Daniel Chu, An Nguyen, Spenser S. Smith, Zuzana Vavrušová, Richard A. Schneider
Generation of clonal male and female mice through CRISPR/Cas9-mediated Y chromosome deletion in embryonic stem cells
Yiren Qin, Bokey Wong, Fuqiang Geng, Liangwen Zhong, Luis F. Parada, Duancheng Wen
Frequent loss-of-heterozygosity in CRISPR-Cas9-edited early human embryos
Gregorio Alanis-Lobato, Jasmin Zohren, Afshan McCarthy, Norah M.E. Fogarty, Nada Kubikova, Emily Hardman, Maria Greco, Dagan Wells, James M.A. Turner, Kathy K. Niakan
FREQUENT GENE CONVERSION IN HUMAN EMBRYOS INDUCED BY DOUBLE STRAND BREAKS
Shoukhrat Mitalipov
Reading frame restoration at the EYS locus, and allele-specific chromosome removal after Cas9 cleavage in human embryos
Michael V. Zuccaro, Jia Xu, Carl Mitchell, Diego Marin, Raymond Zimmerman, Bhavini Rana, Everett Weinstein, Rebeca T. King, Morgan Smith, Stephen H. Tsang, Robin Goland, Maria Jasin, Rogerio Lobo, Nathan Treff, Dieter Egli
Optimized culture of retinal ganglion cells and amacrine cells from adult mice
Yong H Park, Joshua D Snook, Iris Zhuang, Guofu Shen, Benjamin J Frankfort

Application of Airy beam Light sheet microscopy to examine early neurodevelopmental structures in 3D hiPSC-derived human cortical spheroids
Dwaipayan Adhya, George Chennell, James Crowe, Eva P. Valencia-Alarcón, James Seyforth, Neveen Honsy, Marina V. Yasvoina, Robert Forster, Simon Baron-Cohen, Anthony C. Vernon, Deepak. P. Sriavstava
Development of zygotic and germline gene drives in mice
Chandran Pfitzner, James N Hughes, Melissa A White, Michaela Scherer, Sandra G Piltz, Paul Q Thomas
I-KCKT allows dissection-free RNA profiling of adult Drosophila intestinal progenitor cells
Kasun Buddika, Jingjing Xu, Ishara Ariyapala, Nicholas S. Sokol
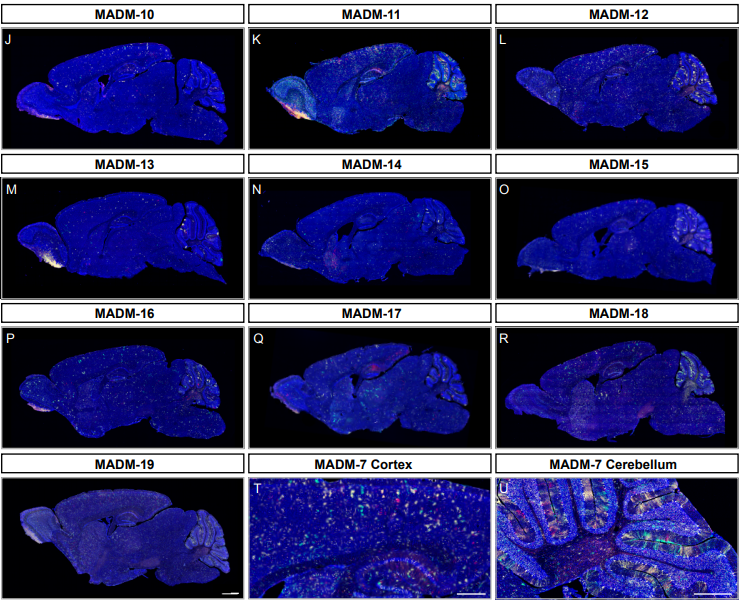
A Genome-wide Library of MADM Mice for Single-Cell Genetic Mosaic Analysis
Ximena Contreras, Amarbayasgalan Davaatseren, Nicole Amberg, Andi H. Hansen, Johanna Sonntag, Lill Andersen, Tina Bernthaler, Anna Heger, Randy Johnson, Lindsay A. Schwarz, Liqun Luo, Thomas Rülicke, Simon Hippenmeyer
In vivo fluorescence imaging with a flat, lensless microscope
Jesse K. Adams, Vivek Boominathan, Sibo Gao, Alex V. Rodriguez, Dong Yan, Caleb Kemere, Ashok Veeraraghavan, Jacob T. Robinson
pHLARE: A Genetically Encoded Ratiometric Lysosome pH Biosensor
Bradley A. Webb, Jessica Cook, Torsten Wittmann, Diane L. Barber
Split-HaloTag® Imaging Assay for in vivo 3D-Microscopy and Subdiffractional Analyses of Protein-Protein Interactions
Rieke Meinen, Jan-Niklas Weber, Andreas Albrecht, Rainer Matis, Maria Behnecke, Cindy Tietge, Stefan Frank, Jutta Schulze, Henrik Buschmann, Peter Jomo Walla, Ralf-R. Mendel, Robert Hänsch, David Kaufholdt
An optogenetic method for interrogating YAP1 and TAZ nuclear-cytoplasmic shuttling
Anna M. Dowbaj, Robert P. Jenkins, Klaus Hahn, Marco Montagner, Erik Sahai
Influence of nanobody binding on fluorescence emission, mobility and organization of GFP-tagged proteins
Falk Schneider, Christian Eggeling, Erdinc Sezgin
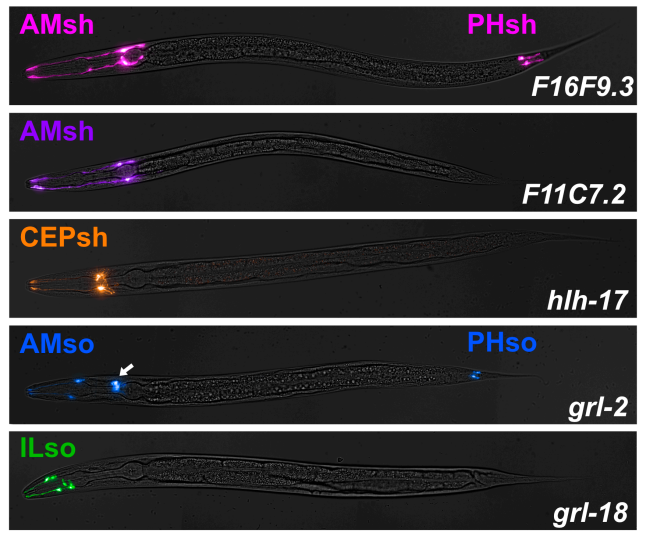
Cell-type-specific promoters for C. elegans glia
Wendy Fung, Leigh Wexler, Maxwell G. Heiman
Research practice & education
International authorship and collaboration across bioRxiv preprints
Richard J. Abdill, Elizabeth M. Adamowicz, Ran Blekhman
Advancing science or advancing careers? Researchers’ opinions on success indicators
Noémie Aubert Bonn, Wim Pinxten
Survey of Australian STEMM Early Career Researchers: job insecurity and questionable research practices are major structural concerns
Katherine Christian, Carolyn Johnstone, Jo-ann Larkins, Wendy Wright, Michael R Doran
20 years of African Neuroscience: Waking a sleeping giant
MB Maina, U Ahmad, HA Ibrahim, SK Hamidu, FE Nasr, AT Salihu, AI Abushouk, M Abdurrazak, MA Awadelkareem, A Amin, A Imam, ID Akinrinade, AH Yakubu, IA Azeez, GM Yunusa, AA Adamu, HB Ibrahim, AM Bukar, AU Yaro, LL Prieto-Godino, T Baden
Only two out of five articles by New Zealand researchers are free-to-access: a multiple API study of access, its impact on open citation advantage, cost of Article Processing Charges (APC), and the potential to increase the proportion of open access
Richard Kenneth Alistair White, Anton Angelo, Deborah Jane Fitchett, Moira Fraser, Luqman Hayes, Jess Howie, Emma Richardson, Bruce Duncan White


 (No Ratings Yet)
(No Ratings Yet)

 (3 votes)
(3 votes)



 (6 votes)
(6 votes)