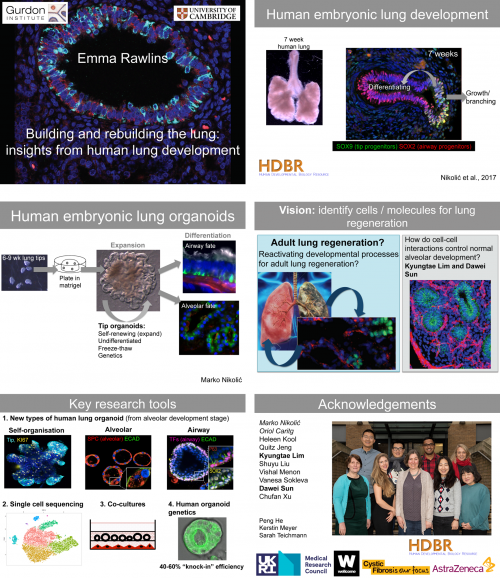
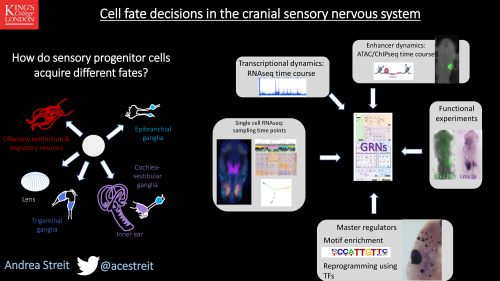
Exhibitors are crucial to fund society conferences like BSDB/Gensoc 2020, and since they did not have the chance to promote themselves in person, here we are showcasing some of them (the post will be added to as more come in, as they understandably have a lot on their plate right now!)

Who are you and what do you do?
Fluidigm develops, manufactures, and markets research products for life science analytical and preparatory systems for use in mass cytometry, high-throughput genomics, and single‑cell genomics applications.
Why were you planning to exhibit at the BSDB/GenSoc meeting this year – how is this community important to you?
Fluidigm offers solutions for people using PCR-based approaches to study SNPs, gene expression changes, copy number variations, and more. At this year’s meeting, we planned to introduce our newest microfluidics product for RNA-Seq Library Preparation on our Juno System. Our solution provides walkaway automation and significant cost savings to researchers who routinely use this application.

If you could get one message across to people registered for the meeting, what would it be?
Our systems are based on proprietary microfluidics technology and are designed to significantly simplify experimental workflow, increase throughput, and reduce costs while providing excellent data quality.

Is there anything else you’d like to share with the BSDB and Genetics Society communities?
You can read about how Dr. Scott Magness at the University of North Carolina School of Medicine has implemented the Advanta RNA-Seq NGS Library Prep Workflow in his laboratory to study intestinal stem cells in 2D model systems, stating that the Advanta system was a “game changer” for their experimental needs.
Read about it here >

How can people sign up to find out more about your company and products/services?
Just go to https://go.fluidigm.com/ContactUs

Who are you and what do you do?
The Royal Society is a charitable organisation that recognises, promotes, and supports excellence in science.

Why were you planning to exhibit at the BSDB/GenSoc meeting this year – how is this community important to you?
We’re regularly attendees and are particularly excited that this year’s meeting joins the Genetics and Developmental Biology community. Our aim is to inform these communities about how we support international collaboration and demonstrate the importance of science to everyone through our publications and wider activities.

If you could get one message across to people registered for the meeting, what would it be?
Our international journals publish high quality science and provide an excellent service to our authors and readers. Open Biology, Proceedings B, Biology Letters, Royal Society Open Science and Philosophical Transactions B – offer publishing options for research, reviews and theme issues within Developmental Biology and Genetics.

Is there anything else you’d like to share with the BSDB and Genetics Society communities?
Reasons to choose our journals include: articles handled by expert biologists; rapid processing; rigorous review; open access, open data, and Registered Reports available. We will also be launching a new article type on Open Biology called ‘Open Questions’ which highlights current advances in an area of biomedical science that is developing quickly and ripe for discovery https://royalsocietypublishing.org/rsob/for-authors#question1
How can people sign up to find out more about your company and products/services?
Go to
https://royalsociety.org/journals/authors/
https://royalsocietypublishing.org/rstb/submit-proposal

Why were you planning to exhibit at the BSDB/GenSoc meeting this year – how is this community important to you?
We’re at this meeting every year – as a publisher of community journals it’s a very important opportunity for us to engage with the developmental biology community and find out how we can better understand and support their needs.
If you could get one message across to people registered for the meeting, what would it be?
We’re a not-for-profit organisation dedicated to supporting biologists in many ways: from providing a smooth publishing journey to connecting people through our community websites; from offering meeting grants and travelling fellowships to organising workshops and meetings.

Is there anything else you’d like to share with the BSDB and Genetics Society communities?
We’ve recently launched the Node Network, a global directory of developmental and stem cell biologists, designed to help find speakers, referees, panel members and potential collaborators. Find out more at https://thenode.biologists.com/network/

How can people sign up to find out more about your company and products/services? (Give us the URL)
Go to
https://www.biologists.com/subscribe
https://thenode.biologists.com/login
 (1 votes)
(1 votes)
 Loading...
Loading...


 (1 votes)
(1 votes)


 (No Ratings Yet)
(No Ratings Yet)