The people behind the papers – Samira Benhamouche-Trouillet, Evan O’Loughlin & Andrea McClatchey
Posted by the Node Interviews, on 21 May 2018
Intrahepatic bile ducts (IHBDs) are epithelial tubular structures that transport bile from the liver to the intestine, but the molecules and mechanisms controlling IHBD morphogenesis have remained largely unclear. A a recent paper in Development reports an investigation into IHBD development and the role the tumour suppressor and cytoskeletal regulator Merlin plays in the process. We caught up first authors Samira Benhamouche-Trouillet and Evan O’Loughlin, and their supervisor Andrea McClatchey, Professor of Pathology at Harvard Medical School and PI at the Massachusetts General Hospital Cancer Centre, to find out more.
Andrea, can you give us your scientific biography and the questions your lab is trying to answer?
AMcC I have always been drawn to scientific discovery and decided early in my training that to apply that to discovering causes and cures for human disease was the most fulfilling thing I could imagine. I completed my PhD training in human genetics in the laboratory of Dr. James Gusella at Mass General Hospital (MGH)/Harvard Medical School (HMS) when DNA sequencing was done by hand and sequencing the human genome was just beginning. During that time, I realized that I was particularly lured by spatial patterns in science – for example, in DNA and protein sequences, the layout of genes on chromosomes or the three-dimensional topography of folded proteins. When I finished, I took on the challenge of cancer research in the new laboratory of Dr. Tyler Jacks at MIT. I decided to focus on the neurofibromatosis type 2 (NF2) tumor suppressor, merlin, that had just been identified by Dr. Gusella’s laboratory. I was fascinated by the fact that merlin was unique among known tumor suppressors as a member of a family of membrane:cytoskeleton linking proteins (the ERMs; ezrin, radixin and moesin). I imagined how these proteins might organize topographical patterns on the surface of cells and tissues and how that could be important for preventing inappropriate cell behavior. In studies that began then and carried forward to my own laboratory at MGH/HMS, we have come to appreciate the important functions that merlin and the ERMs have in organizing the interface between the plasma membrane and underlying cortical cytoskeleton during tissue morphogenesis and tumorigenesis. My lab is trying to understand the molecular and cellular mechanisms by which merlin/ERMs organize this critical cellular compartment in normal tissues and how it is defective in tumors, with the ultimate goal of harnessing that information to develop new therapies for NF2-mutant and other cancers. This work by Samira and Evan is an important example of that strategy. We believe this work sets the stage for understanding how aspects of biliary morphogenesis, and NF2-deficiency specifically, contribute to biliary tumorigenesis, and for developing ways to interfere with those processes therapeutically.
Samira and Evan: how did you come to work in Andrea’s lab and be involved with this project?
SB-T I met Andi, in 2006, after a seminar she gave at the Curie Institute in Paris about the role of ezrin in intestinal development and morphogenesis and its implications in diseases. I was very intrigued by her first slide mentioning the role of Nf2/merlin in liver cancers – both hepato- and cholangio-cellular carcinomas. Andi’s work was extremely elegant and I understood quickly that I wanted to work with her in understanding the role of Nf2/merlin and the membrane-cytoskeletal interface in controlling cell polarity and cell fate/plasticity. When I joined the lab in 2008, I had the chance to learn from Marcello Curto, a postdoctoral fellow in the lab who had developed the mouse model of Nf2 loss in the liver (Alb-cre;Nf2 lox/lox). We handled together the phenotypic characterization of the model and showed that Nf2 loss leads to neoductulogenesis in adult mice, and eventually liver cancer (Benhamouche, Curto, et al 2010). I further wanted to understand the origin of the defects in Nf2 null livers and took advantage of recent data from Dr. Lemaigre’s group showing the role of Sox9+ ductal plate cells as the cell of origin for adult liver progenitor cells, biliary cells and periportal hepatocytes (Antoniou, et al 2009, Carpentier et al, 2011). This led us to study how merlin controls bile duct development.
EO’L I joined the McClatchey lab because I have a longstanding interest in understanding development alongside the perspective of cell biology. How cells undergo dramatic, choreographed rearrangements with respect to each other to shape complex tissues like a branched network of tubes is a fascinating question for me. I think it’s an issue that requires us to both examine the behavior of individual cells (For example, how are they oriented? What molecules are required for cells to appropriately “sense” their neighbors?), as well as the organization of cell collectives over time. When I arrived in Andi’s lab, Samira and a former postdoctoral fellow, Marcello Curto, had done beautiful work demonstrating that NF2 is required to restrict cell fate in the liver, and that its loss can lead to liver cancer. More recently, Samira, wondering if this cell fate change was a developmental phenomenon, investigated the origins of this phenotype, describing how biliary morphogenesis goes awry in the Nf2-mutant embryonic liver. When I joined the lab after Samira left, I was immediately drawn to this project mainly because liver development just seemed so remarkable to me. How do cells go from existing as an undifferentiated “mass” of hepatoblasts to building tubes within a tissue, that have to be the correct proportions and align properly with adjacent tubes?
What was known about biliary tube morphogenesis before your work?
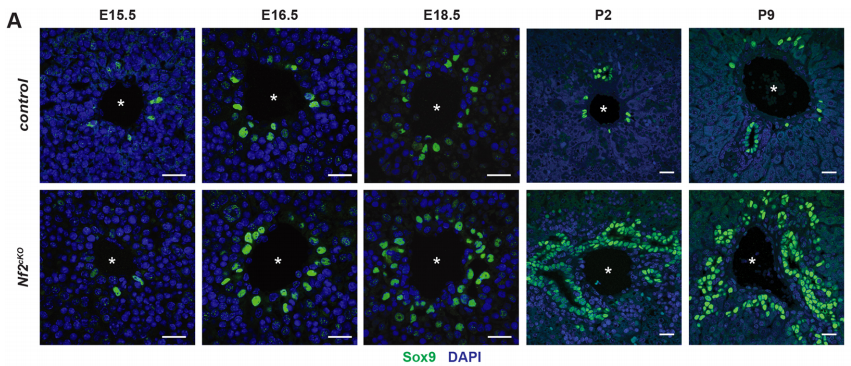
SB-T Not much was really known regarding morphogenesis. Implications of signalling pathways such as Notch and Wnt/beta-catenin had been reported but lack of good tracers made it difficult to image the in vivo process of bile duct morphogenesis. The use of lineage reporter mice, improvement of microscopy techniques and the discovery of Sox9 cells as the earliest event of bile duct development with the formation of asymmetric primitive ductular structure (Antoniou et al, 2009, Zong et al, 2009), brought up new insights for me to ask whether merlin was involved in limiting the distribution of ERM apical complexes and Sox9+ ductal plate cell fate.
Can you give us the key results of the paper in a paragraph?
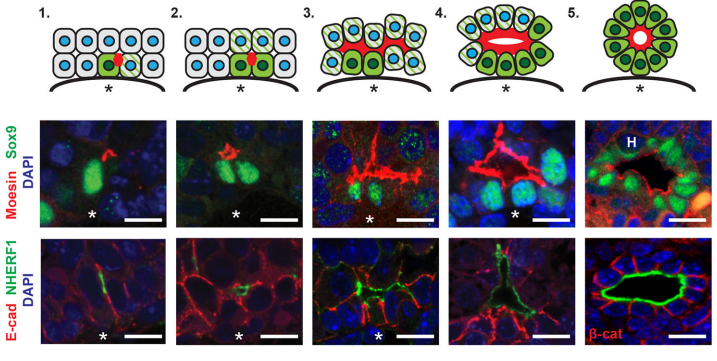
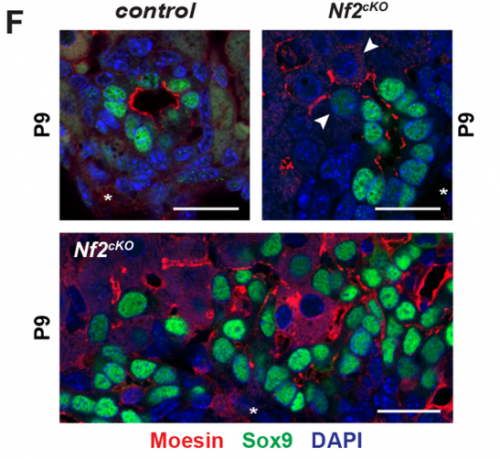
AMcC We wanted to understand how Nf2 mutation causes massive biliary neoductulogenesis and tumorigenesis in the mouse. In tracing the origin of the phenotype back to the developing liver, we realized that the defect begins at a remarkable but poorly understood stage of biliary morphogenesis, and if we were to understand how Nf2-deficiency caused it we would need to understand how the earliest stages of normal biliary morphogenesis proceeds. Samira and Evan discovered that the formation of a ‘tree’ of biliary tubes within a bed of hepatoblasts proceeds via a self-organizing process that is initiated by a single cell that adopts a biliary fate, becomes polarized and creates a tiny apical microlumen at a boundary with an adjacent undifferentiated hepatoblast. Adjacent cells are then recruited to the developing biliary tube via a process involving cell-cell communication, lumen expansion and apical constriction. Importantly the formation of the biliary tree is entirely inductive and occurs without proliferation. Samira and Evan’s studies suggest that the recruitment of cells into the developing tubes ceases for mechanical reasons – when apical constriction limits lumen expansion. In the absence of merlin this recruitment continues unabated, yielding an abundance of disorganized biliary structures without proliferation.
It’s quite remarkable that a complex morphogenetic can be initiated by a single cell. Do you have any idea how this one cell convinces its neighbors to join in?
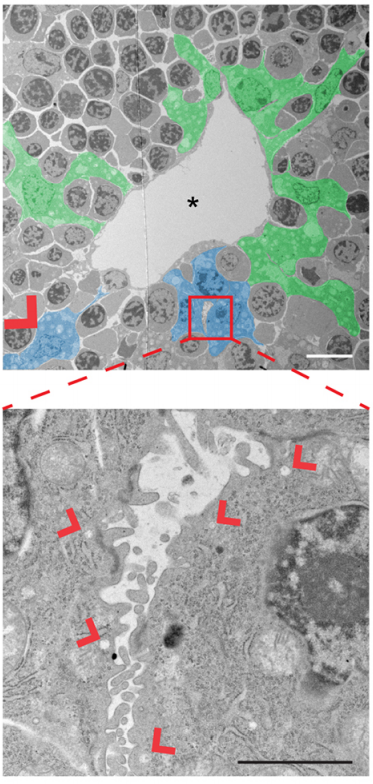
EO’L Yes, we were really struck by the observation that we consistently observed single biliary cells with luminal “puncta” on their lateral boundary, and this appears to be the “starting point” for generating whole tubes. I have some theories and we would definitely like to investigate these hypotheses further in future studies. I think it’s important to consider that deposition of luminal material on the boundary of a cell means that cell’s neighbor now also has an apical domain. And, after this event, the neighbor seems to also turn on Sox9, a biliary marker. So, we believe that this initial polarization cue may be instructive to the fate of the neighboring cell. We don’t yet know what that signal is, although it has been shown in other model systems like the zebrafish lateral line primordium that the acquisition of luminal architecture can serve to spatially restrict growth factor signaling, which can then feedback and promote cell fate (Durdu et al, 2014). Especially given that our lab has shown that a key function of NF2 is to restrict the membrane distribution of EGFR, we speculate that having spatially-restricted RTK signaling within this nascent lumen may promote the biliary differentiation of cells that contact the lumen. Thus, as the lumen itself grows, more and more biliary cells are incorporated.
What do you think the most important implications of your work are for liver disease and regeneration?
EO’L I think that a lot of disease processes probably involve the inappropriate activation of developmental programs in mature tissues. In particular, many liver diseases such as cholangiocarcinoma feature “neoductulogenesis”, the rampant formation of new ducts. I think that a mechanistic understanding of this phenomenon will require both deciphering the specific signals that trigger duct formation and the factors that serve to limit the expansion of these tubes during development. I think our work provides novel insight into how biliary tubes “get started” and how a tumor suppressor such as NF2 (which has recently been found to be mutated in a subset of human biliary cancers, by the way) can halt the growth of these ductular lesions.
SB-T This work highlights the importance of coordinating lumen formation and cell fate for liver homeostasis. I hope that this work will open new insights to approach liver cell plasticity in liver diseases but also how to control cell behavior for regenerative medicine.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
SB-T I was really excited when I observed for the first time that defects in the Nf2-deficient livers were proliferation independent and associated with excessive lumen expansions. This suggested that the latter could be responsible for the over-recruitment and over-conversion of Sox9 positive cells at sites of de novo tubulogenesis.
EO’L I think a big one for me was the observation of single polarized cells, which we touched on above. A second was when I started measuring the sizes of the apical and basal surfaces in the bile ducts of livers lacking NF2. I knew the lumens were much too large, but I wasn’t expecting so many cells to have such a distorted apical:basal ratio – often the apical surface is several-fold larger than the basal side! It’s as if, in the absence of NF2, the apical surface just “springs open”!
And what about the flipside: any moments of frustration or despair?
SB-T Research is a journey with ups and downs but I was always confident with the results. Despite some frustration concerning the lack of in vitro tools to test our model and bring further mechanistic explanations, we are showing for the first time how bile duct tubulogenesis proceeds and how this impacts our understanding of liver cell differentiation.
EO’L I think I initially didn’t appreciate the full complexity of liver development and so Andi and I spent a lot of time going back and forth about how to make sure we were capturing everything that goes on. It’s especially hard when you’re looking at fixed tissues, which can only be a “snapshot” in time. For example, it’s evident that not all portal veins are created equal: larger ones, closer to the “base” of the tree, are sites of biliary differentiation well before veins on the periphery. Also, not everything happens synchronously – so you can have a portal vein, for example, with early, middle, and late stages of biliary differentiation happening simultaneously. Finally, it’s important to recognize that everything we describe happens in three-dimensions. Thanks to our wonderful collaborator Bill Polacheck in Chris Chen’s lab, we tried to provide some insight into how the initial shapes of the lumens differ in 3D in the mutant, but how these lumens expand along the z-axis later in development, and anastomose, are outstanding questions.
What next for you two after this paper?
EO’L I am currently interested in the three-dimensional culture of liver cells, and in using these methods to answer some of the more molecular questions that the current study left unanswered – such as whether there are specific signaling pathways, downstream of NF2, that promote biliary expansion. I have found ways, for example, to coax primary hepatoblasts into making tubes in vitro. I believe 3D culture can complement both traditional cell culture, which has the disadvantage of being a highly artificial environment for cells, and in vivo work, which can be expensive and time-consuming.
SB-T After this tremendous training in Andi’s lab, I came back to France to develop my own group. We have uncovered the physiological role of an AAA+ ATPase protein called Reptin in liver metabolism and homeostasis (Javary et al, 2017). Now I am currently developing a project to understand cell heterogeneity in the liver and how this can affect liver regeneration.
And where will this work take the McClatchey lab?
AMcC We are already focused on follow-up studies in several areas. First, with the help of our bioengineering colleagues, we are working to develop an in vitro model that ultimately recapitulates the self-organizing process of biliary tube formation from within a mass of hepatoblasts. This will allow us to dissect the molecular mechanisms by which this process normally occurs as well as those that interfere with it. This has important implications for tissue engineering, liver regeneration and biliary tumorigenesis. Second, we are working to understand these cellular and molecular mechanisms may be aberrantly re-enacted to drive or contribute to tumorigenesis. For example, how do the neoductular patterns that are seen in human biliary tumors arise? Can an aberrant biliary cell recruit normal neighbors into a developing tumor, creating an important source of heterogeneity? How does aberrant proliferation ultimately arise? How do known mutational drivers of biliary tumorigenesis affect this self-organizing process? Can we interfere with specific aspects of this process as a therapeutic strategy? Finally, how can this information inform our understanding of other types of Nf2-mutant tumors?
Finally, let’s move outside the lab – what do you like to do in your spare time?
SB-T Outside the lab, I love spending time with my family. We travel and visit places all around; I enjoy hiking and reading thriller and history books.
EO’L I grew up in the beautiful mountains of Boulder, Colorado, so getting outdoors and into nature is sort of my lifeblood. It’s definitely a bit harder in Boston, but I try to find time to go to nearby parks and nature reservations for a walk. I especially enjoy birdwatching. I actually think watching birds is a bit like developmental biology, making careful observations and looking for patterns, and it also satisfies my curiosity about the world around me.
AMcC Believe it or not I love to curl up and read a cool science paper with a little bit of hard-to-come-by spare time, and to let my mind be free to consider scientific connections, with our own work or maybe with something else I read. Outside of science I am a big music buff (particularly blues, jazz and rock) and a big sports fan, and I have one or the other on the radio a lot of the time. I also grew up hiking and backpacking in the mountains (of New England), and like nothing better than to be on a nice long hike, maybe with my son and my dog Fenway, particularly in the Maine wilderness where my family has a log cabin.
Proliferation-independent role of NF2 (merlin) in limiting biliary morphogenesis
Samira Benhamouche-Trouillet, Evan O’Loughlin, Ching-Hui Liu, William Polacheck, Julien Fitamant, Mary McKee, Nabeel El-Bardeesy, Christopher S. Chen, Andrea I. McClatchey
Development 2018 145: dev162123 doi: 10.1242/dev.162123
This is #41 in our interview series. Browse the archive here.


 (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)









 I’m extremely happy to be acting as the new Graduate Representative for BSDB, following
I’m extremely happy to be acting as the new Graduate Representative for BSDB, following  Having completed my PhD in the field of vertebrate somitogenesis in the lab of
Having completed my PhD in the field of vertebrate somitogenesis in the lab of  Tanya is Professor of Developmental Biology at the University of Sheffield, where she is a member of the Bateson Centre and Department of Biomedical Science [
Tanya is Professor of Developmental Biology at the University of Sheffield, where she is a member of the Bateson Centre and Department of Biomedical Science [ Shankar is Professor of Developmental Biology and a Wellcome Senior Investigator in the Department of Physiology Anatomy and Genetics at the University of Oxford [
Shankar is Professor of Developmental Biology and a Wellcome Senior Investigator in the Department of Physiology Anatomy and Genetics at the University of Oxford [ Jens is a Sir Henry Dale fellow at the School of Life Sciences at the University of Dundee running his lab in the division of Cell and Developmental Biology [
Jens is a Sir Henry Dale fellow at the School of Life Sciences at the University of Dundee running his lab in the division of Cell and Developmental Biology [
