November in preprints
Posted by the Node, on 5 December 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. Let us know if we missed anything.
This month in preprint news – here at The Company of Biologists, set just outside Cambridge in the UK, we’re hiring a new ‘Preprints Community Manager’. You can find out more here. It’s a great opportunity to get in to science  communication and publishing, and of course preprints!
communication and publishing, and of course preprints!
And this month in preprints, we found platypuses and pigeons, xenotransplants and reoriented axes, CRISPR tools and liquid droplets. The preprints were hosted on bioRxiv, PeerJ, and arXiv. Use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice
Why not…
Developmental biology
| Patterning & signalling
Reorientation of the primary body axis by ectopic embryonic cWnt signaling. Mark Q Martindale, Naveen Wijesena
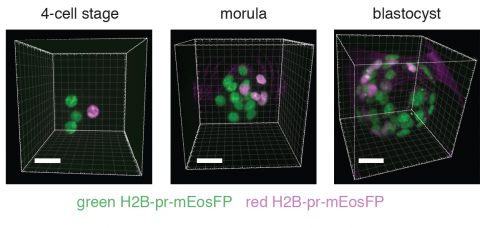
High fidelity lineage tracing in mouse pre-implantation embryos using primed conversion of photoconvertible proteins. Maaike Welling, Manuel A. Mohr, Aaron Ponti, Lluc R. Sabater, Andrea Boni, Prisca Liberali, Pawel Pelczar, Periklis Pantazis
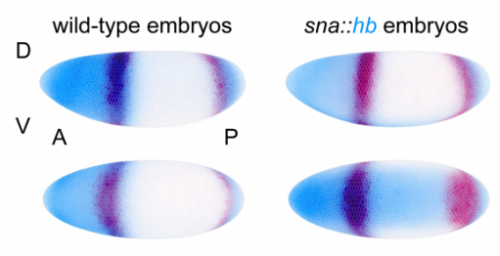
Caudal counter-represses Hunchback to regulate even-skipped stripe 2 expression in Drosophila embryos. Ben J Vincent, Max V Staller, Francheska Lopez-Rivera, Meghan DJ Bragdon, Zeba Wunderlich, Javier Estrada, Angela H DePace
A damped oscillator imposes temporal order on posterior gap gene expression in Drosophila. Berta Verd, Erik Clark, Karl R. Wotton, Hilde Janssens, Eva Jimenez-Guri, Anton Crombach, Johannes Jaeger
From pioneer to repressor: Bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Daria Gavriouchkina, Ruth M Williams, Martyna Lukoseviciute, Tatiana Hochgreb-Hagele, Upeka Senanayake, Vanessa Chong-Morrison, Supat Thongjuea, Emmanouela Repapi, Adam Mead, Tatjana Sauka-Spengler
Embryonic circular RNAs at tandem duplicated genes in zebrafish present new paradigm in gene expression. Vanessa Chong-Morrison, Tatjana Sauka-Spengler
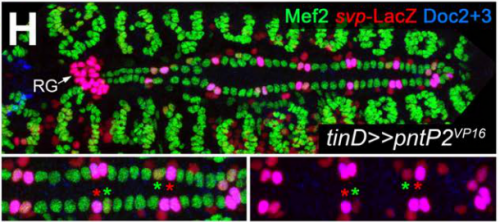
Diversification of heart progenitor cells by EGF signaling and differential modulation of ETS protein activity. Benjamin Schwarz, Dominik Hollfelder, Katharina Scharf, Leonie Hartmann, Ingolf Reim
Single cell RNA-seq and ATAC-seq indicate critical roles of Isl1 and Nkx2-5 for cardiac progenitor cell transition states and lineage settlement. Guangshuai Jia, Jens Preussner, Stefan Guenther, Xuejun Yuan, Michail Yekelchyk, Carsten Kuenne, Mario Looso, Yonggang Zhou, Thomas Braun
Early Subset Of Cerebellar Nuclei Neurons Derived From Mesencephalon In Mice. Maryam Rahimi-Balaei, Xiaodan Jiao, Fiona Parkinson, Behzad Yeganeh, Hassan Marzban
Elevated and sustained reactive oxygen species levels facilitate mesoderm formation during early Xenopus development. Yue Han, Yaoyao Chen, Nick R Love, Shoko Ishibashi, Enrique Amaya
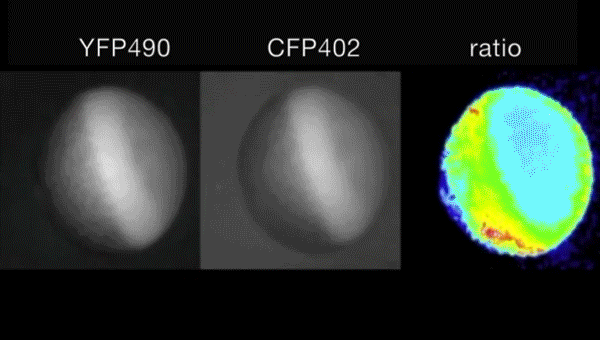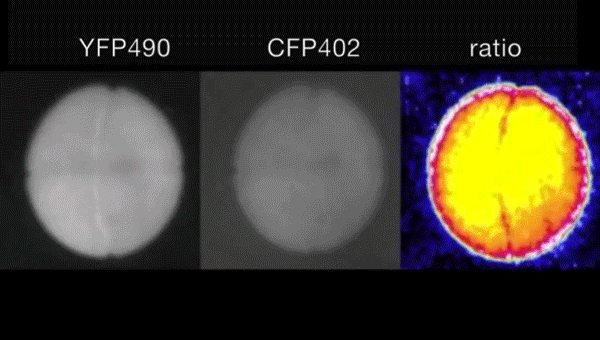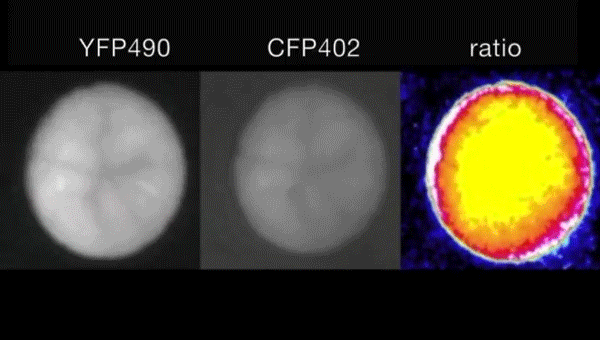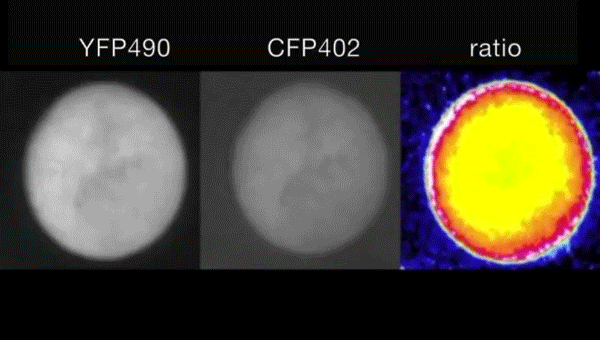
Ca2+-induced mitochondrial ROS regulate the early embryonic cell cycle. Yue Han, Shoko Ishibashi, Javier Iglesias-Gonzalez, Yaoyao Chen, Nick R Love, Enrique Amaya
Proteomics of phosphorylation and protein dynamics during fertilization and meiotic exit in the Xenopus egg. Marc Stuart Presler, Elizabeth Van Itallie, Allon M. Klein, Ryan Kunz, Peg Coughlin, Leonid Peshkin, Steven Gygi, Martin Wühr, Marc Kirschner
Maternal proteins that are phospho-regulated upon egg activation include crucial factors for oogenesis, egg activation and embryogenesis in Drosophila melanogaster. Zijing Zhang, Amber R. Krauchunas, Stephanie Huang, Mariana F. Wolfner
Endodermal differentiation is reconstructed by coordination of two parallel signaling systems derived from the stele in roots. Pengxue Li, Qiaozhi Yu, Chunmiao Xu, Xu Gu, Shilian Qi, Hong Wang, Fenglin Zhong, Shuang Wu
Minimum requirements for reprogramming and maintaining cell fate in the Arabidopsis root. Colleen Drapek, Erin E. Sparks, Peter Marhavy, Tonni G. Andersen, Jessica H. Hennacy, Niko Geldner, Philip N. Benfey
LRX Proteins play a crucial role in pollen grain and pollen tube cell wall development. Tohnyui Ndinyanka Fabrice, Hannes Vogler, Christian Draeger, Gautam Munglani, Shibu Gupta, Aline Galatea Herger, Paul Knox, Ueli Grossniklaus, Christoph Ringli
Hominin-specific NOTCH2 paralogs expand human cortical neurogenesis through regulation of Delta/Notch interactions. Ikuo K Suzuki, David Gacquer, Roxane Van Heurck, Devesh Kumar, Marta Wojno, Angeline Bilheu, Adele Herpoel, Julian Cheron, Franck Polleux, Vincent Detours, Pierre Vanderhaeghen
Human-specific NOTCH-like genes in a region linked to neurodevelopmental disorders affect cortical neurogenesis. Ian T Fiddes, Gerrald A Lodewijk, Meghan M Mooring, Colleen M Bosworth, Adam D Ewing, Gary L Mantalas, Adam M Novak, Anouk van den Bout, Alex Bishara, Jimi L Rosenkrantz, Ryan Lorig-Roach, Andrew R Field, Maximillian Haeussler, Lotte Russo, Aparna Bhaduri, Tomasz J Nowakowski, Alex A Pollen, Max L Dougherth, Xander Nuttle, Marie-Claude Addor, Simon Zwolinski, Sol Katzman, Arnold Kreigstein, Evan E Eichler, Sofie R Salama, Frank MJ Jacobs, David Haussler
Skeletal muscle fibro-adipogenic progenitors of dystrophic mice are insensitive to NOTCH-dependent regulation of adipogenesis. Milica Marinkovic, Francesca Sacco, Filomena Spada, Lucia Lisa Petrilli, Claudia Fuoco, Elisa Micarelli, Theodora Pavlidou, Luisa Castagnoli, Matthias Mann, Cesare Gargioli, Gianni Cesareni
NOTCH-mediated non-cell autonomous regulation of chromatin structure during senescence. Aled J Parry, Matthew Hoare, Dóra Bihary, Robert Hänsel-Hertsch, Stephen Smith, Kosuke Tomimatsu, Shankar Balasubramanian, Hiroshi Kimura, Shamith Samarajiwa, Masashi Narita
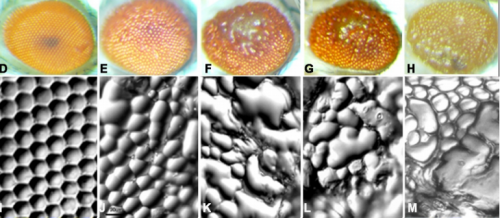
Altered hsrω lncRNA levels in activated Ras background further enhance Ras activity in Drosophila eye and induces more R7 photoreceptors. Subhash C Lakhotia, Mukulika Ray
An atlas of Caenorhabditis elegans chemoreceptor expression. Berta Vidal, Ulkar Aghayeva, Haosheng Sun, Chen Wang, Lori Glenwinkel, Emily Bayer, Oliver Hobert
Bidirectional Cycling Dynamics of Living Neuronal Networks in Vitro. Arseniy Gladkov, Oleg Grinchuk, Yana Pigareva, Irina Mukhina, Victor Kazantsev, Alexey Pimashkin
Loss of Wt1 in the murine spinal cord alters interneuron composition and locomotion. Danny Schnerwitzki, Sharn Perry, Anna Ivanova, Fabio Viegas Caixeta, Paul Cramer, Sven Guenther, Kathrin Weber, Atieh Tafreshiha, Lore Becker, Ingrid L. Vargas Panesso, Thomas Klopstock, Martin Hrabe de Angelis, Manuela Schmidt, Klas Kullander, Christoph Englert
Smad9 is a key player of follicular selection in goose via keeping the balance of LHR transcription. Daolun Yu, Fanghui Chen, Li Zhang, Hejian Wang, Jie Chen, Zongmeng Zhang, Jie Li, Chaofeng Xing, Honglin Li, Jun Li, Yafei Cai
Widespread dynamic and pleiotropic expression of the melanocortin-1-receptor (MC1R) system is conserved across chick, mouse and human embryonic development. Anna Thomas, Pauline Heux, Chloe Santos, Wisenave Arulvasan, Nita Solanky, Magalie E. Carey, Dianne Gerrelli, Veronica A. Kinsler, Heather C. Etchevers
| Morphogenesis & mechanics
Size-reduced embryos reveal a gradient scaling based mechanism for zebrafish somite formation. Kana Ishimatsu, Tom W Hiscock, Zach M Collins, Dini Wahyu Kartika Sari, Kenny Lischer, David L. Richmond, Yasumasa Bessho, Takaaki Matsui, Sean G Megason
Spatio-temporal elasticity patterns in extracellular matrix during Hydra morphogenesis. Mariam Veschgini, Hendrik O Petersen, Wasim Abuillan, Fernanda F Rossetti, Suat Özbek, Manfred Burghammer, Thomas W Holstein, Motomu Tanaka
Selective Filopodia Adhesion Ensures Robust Cell Matching in the Drosophila Heart. ShaoBo Zhang, Christopher Amourda, Timothy Saunders
Junction-based lamellipodia drive endothelial cell rearrangements in vivo via a VE-cadherin/F-actin based oscillatory ratchet mechanism. Ilkka Paatero, Loic Sauteur, MInkyoung Lee, Anne Karine Lagendijk, Daniel Heutschi, Dimitri Bieli, Benjamin M. Hogan, Markus Affolter, Heinz-Georg Belting
Actomyosin contractility modulates Wnt signaling through adherens junction stability. Eric T. Hall, Elizabeth A. Hoesing, Endre D. Sinkovics, Esther M. Verheyen
PI(4,5)P2 forms dynamic cortical structures and directs actin distribution and cell polarity in C. elegans embryos. Melina J. Scholze, Kevin S. Barbieux, Alessandro De Simone, Mathilde Boumasmoud, Camille C. N. Suess, Ruijia Wang, Pierre Gonczy
Atf3 links loss of epithelial polarity to defects in cell differentiation and cytoarchitecture. Colin D. Donohoe, Andreia Correia, Gabor Csordas, Marek Jindra, Corinna Klein, Bianca Habermann, Mirka Uhlirova
Cerebellar granule cell precursors can extend processes, undergo short migratory movements and express postmitotic markers before mitosis in the chick EGL. Michalina Hanzel, Richard JT Wingate
Axons from the trigeminal ganglia are the earliest afferent projections to the mouse cerebellum. Hassan Marzban, Maryam Rahimi Balaei, Richard Hawkes
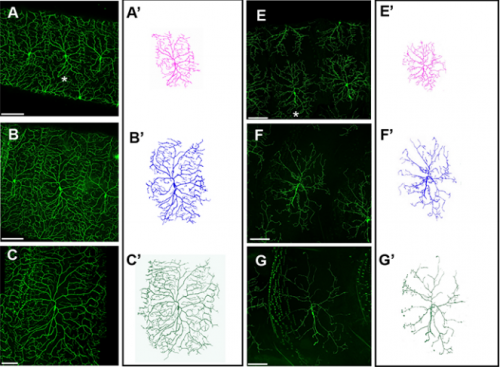
Formin3 regulates dendritic architecture via microtubule stabilization and is required for somatosensory nociceptive behavior. Ravi Das, Jamin M Letcher, Jenna M Harris, Istvan Foldi, Sumit Nanda, Hansley M Bobo, József Mihály, Giorgio A Ascoli, Daniel N Cox
Effects of mutating α-tubulin lysine 40 on sensory dendrite development. Brian Jenkins, Harriet A. J. Saunders, Helena L. Record, Dena M. Johnson-Schlitz, Jill Wildonger
Anisotropic Cellular Mechanoresponse for Radial Size Maintenance of Developing Epithelial Tubes. Tsuyoshi Hirashima, Taiji Adachi
Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis. Kenzo Ivanovitch, Susana Temino, Miguel Torres
| Genes & genomes
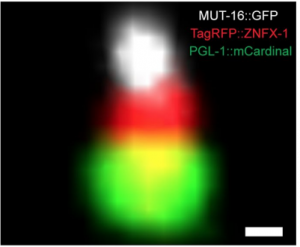
Transgenerational Epigenetic Inheritance Factors Localize to Spatially and Temporally Ordered Liquid Droplet Assemblages. Gang Wan, Brandon Fields, George Spracklin, Carolyn Phillips, Scott Kennedy
Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. Andreas Sagner, Zachary Gaber, Julien Delile, Jennifer H. Kong, David L. Rousso, Caroline A. Pearson, Steven E. Weicksel, Manuela Melchionda, Neda Mousavy Gharavy, James Briscoe, Bennett Novitch
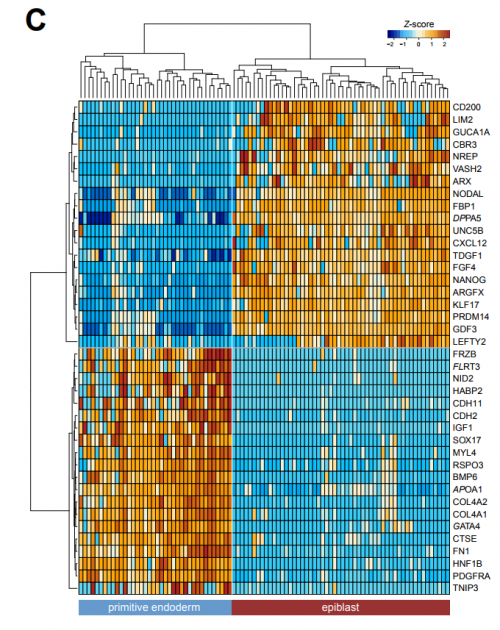
Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human preimplantation epiblast. Giuliano G Stirparo, Thorsten Boroviak, Ge Guo, Jennifer Nichols, Austin Smith, Paul Bertone
Allele-specific control of replication timing and genome organization during development. Juan Carlos Rivera-Mulia, Andrew Dimond, Daniel Vera, Claudia Trevilla-Garcia, Takayo Sasaki, Jared Zimmerman, Catherine Dupont, Joost Gribnau, Peter Fraser, David M Gilbert
The conserved LEM-3/Ankle1 nuclease is involved in the combinatorial regulation of meiotic recombination repair and chromosome segregation in Caenorhabditis elegans. YE HONG, Maria Velkova, Nicola Silva, Marlene Jagut, Viktor Scheidt, Karim Labib, Verena Jantsch, Anton Gartner
Re-arrangements in the cytoplasmic distribution of small RNAs following the maternal-to-zygotic transition in Drosophila embryos. Mehmet Ilyas Cosacak, Hatice Yigit, Bunyamin Akgul
Smg5 is required for multiple nonsense-mediated mRNA decay pathways in Drosophila. Jonathan O. Nelson, Dominique Foerster, Kimberly Frizzell, Stefan Luschnig, Mark M. Metzstein
Quantitative RNAseq Meta Analysis Of Alternative Exon Usage In C. elegans. Nicolas J. Tourasse, Jonathan R. M. Millet, Denis Dupuy
Dynamic reorganization of nuclear architecture during human cardiogenesis. Paul A Fields, Vijay Ramani, Giancarlo Bonora, Galip Gurkan Yardimci, Alessandro Bertero, Hans Reinecke, Lil Pabon, William S Noble, Jay Shendure, Charles Murry
Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. Erin E. Terry, Xiping Zhang, Christy Hoffmann, Laura D. Hughes, Scott A. Lewis, Jiajia Li, Lance Riley, Nicholas F. Lahens, Ming Gong, Francisco Andrade, Karyn A. Esser, Michael E. Hughes
Deciphering cis-regulatory logic with 100 million synthetic promoters. Carl de Boer, Ronen Sadeh, Nir Friedman, Aviv Regev
Spatially varying cis-regulatory divergence in Drosophila embryos elucidates cis-regulatory logic. Peter A. Combs, Hunter B. Fraser
Determinants of Transcription Initiation Directionality in Metazoans. Mahmoud M Ibrahim, Aslihan Karabacak, Alexander Glahs, Ena Kolundzic, Antje Hirsekorn, Alexa Carda, Baris Tursun, Robert P Zinzen, Scott A Lacadie, Uwe Ohler
Transcription start site analysis reveals widespread divergent transcription in D. melanogaster and core promoter encoded enhancer activities. Sarah Rennie, Maria Dalby, Marta Lloret-Llinares, Stylianos Bakoulis, Christian Dalager Vaagenso, Torben Heick Jensen, Robin Andersson
Higher-order inter-chromosomal hubs shape 3-dimensional genome organization in the nucleus. Sofia A Quinodoz, Noah Ollikainen, Barbara Tabak, Ali Palla, Jan Marten Schmidt, Elizabeth Detmar, Mason Lai, Alexander Shishkin, Prashant Bhat, Vickie Trinh, Erik Aznauryan, Pamela Russell, Christine Cheng, Marko Jovanovic, Amy Chow, Patrick McDonel, Manuel Garber, Mitchell Guttman
CHRAC/ACF Contribute to the Repressive Ground State of Chromatin. Alessandro Scacchetti, Laura Brueckner, Dhawal Jain, Tamas Schauer, Xu Zhang, Frank Schnorrer, Bas van Steensel, Tobias Straub, Peter B. Becker
Strong binding activity of few transcription factors is a major determinant of open chromatin. Bei Wei, Arttu Jolma, Biswajyoti Sahu, Lukas M. Orre, Fan Zhong, Fangjie Zhu, Teemu Kivioja, Inderpreet Kaur Sur, Janne Lehtio, Minna Taipale, Jussi Taipale
Chromatin-release of the long ncRNA A-ROD is required for transcriptional activation of its target gene DKK1. Evgenia Ntini, Julia Liz, Jose M Muino, Annalisa Marsico, Ulf Andersson Oerom
LEM-3 is a midbody-tethered DNA nuclease that resolves chromatin bridges during cytokinesis. YE HONG, Remi Sonneville, Bin Wang, Viktor Scheidt, Bettina Meier, Alexander Woglar, Sarah Demetriou, Karim Labib, Verena Jantsch, Anton Gartner
Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Kelsey A. Maher, Marko Bajic, Kaisa Kajala, Mauricio Reynoso, Germain Pauluzzi, Donnelly A. West, Kristina Zumstein, Margaret Woodhouse, Kerry Bubb, Mike Dorrity, Christine Queitsch, Julia Bailey-Serres, Neelima Sinha, Siobhan M. Brady, Roger B. Deal
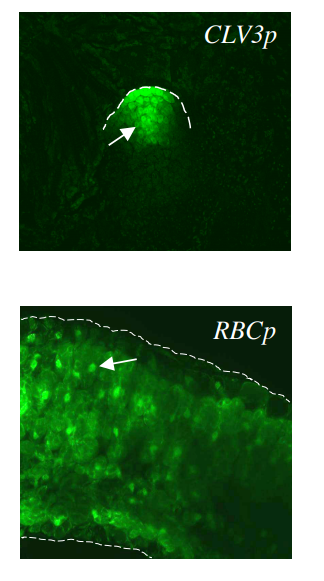
Chromatin accessibility changes between Arabidopsis stem cells and mesophyll cells illuminate cell type-specific transcription factor networks. Paja Sijacic, Marko Bajic, Elizabeth C. McKinney, Richard B. Meagher, Roger B. Deal
Distinct types of short open reading frames are translated in plant cells. Igor Fesenko, Ilya Kirov, Andrey Kniazev, Regina Khazigaleeva, Vassili Lazarev, Daria Kharlampieva, Ekaterina Grafskaia, Viktor Zgoda, Ivan Butenko, Georgy Arapidi, Anna Mamaeva, Vadim Ivanov, Vadim Govorun
Distinct stimulatory mechanisms regulate the catalytic activity of Polycomb Repressive Complex 2 (PRC2). Chul-Hwan Lee, Marlene Holder, Daniel Grau, Ricardo Saldana-Meyer, Rais Ahmad Ganai, Jenny Zhang, Miao Wang, Marc-Werner Dobenecker, Danny Reinberg, Karim-Jean Armache
Functional redundancy of variant and canonical histone H3 lysine 9 modification in Drosophila. Taylor J. R. Penke, Daniel J. McKay, Brian D. Strahl, A. Gregory Matera, Robert J. Duronio
The function of the prereplicative complex and SCF complex components during Drosophila wing development. Hidetsugu Kohzaki
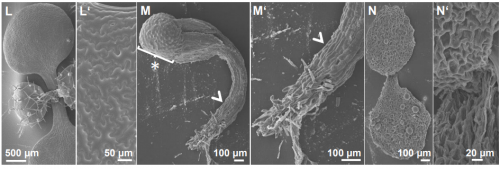
PWO1 interacts with PcG proteins and histones to regulate Arabidopsis flowering and development. Mareike L Hohenstatt, Pawel Mikulski, Olga Komarynets, Constanze Klose, Ina Kycia, Albert Jeltsch, Sara Farrona, Daniel Schubert
PWWP INTERACTOR OF POLYCOMBS (PWO1) links PcG-mediated gene repression to the nuclear lamina in Arabidopsis. Pawel Mikulski, Mareike L Hohenstatt, Sara Farrona, Cezary Smaczniak, Kerstin Kaufmann, Gerco Angenent, Daniel Schubert
The HoxD Cluster is a Dynamic and Resilient TAD Boundary Controling the Segregation of Antagonistic Regulatory Landscapes. Eddie Rodriguez-Carballo, Lucille Lopez-Delisle, Ye Zhan, Pierre Fabre, Leonardo Beccari, Imane El-Idrissi, Thi Hahn Nguyen Huynh, Hakan Ozadam, Job Dekker, Denis Duboule
Cohesin facilitates zygotic genome activation in zebrafish. Michael Meier, Jenny Grant, Amy Dowdle, Amarni Thomas, Jennifer E. Gerton, Philippe Collas, Justin M. O’Sullivan, Julia A. Horsfield
CTCF mediated genome architecture regulates the dosage of mitotically stable mono-allelic expression of autosomal genes. Keerthivasan Raanin Chandradoss, Kuljeet Singh Sandhu
Interdependent and separable functions of C. elegans MRN-C complex members couple formation and repair of meiotic DSBs. Chloe Girard, Baptiste Roelens, Karl A. Zawadzki, Anne M. Villeneuve
LINE-2 transposable elements shape post-transcriptional gene regulation in the human brain. Rebecca Petri, Per Ludvik Brattås, Marie E. Jönsson, Karolina Pircs, Johan Bengzon, Johan Jakobsson
No kissing in the nucleus: Unbiased analysis reveals no evidence of trans chromosomal regulation of mammalian immune development. Timothy M Johanson, Hannah D Coughlan, Aaron TL Lun, Naiara G Bediaga, Gaetano Naselli, Alexandra L Garnham, Leonard C Harrison, Gordon K Smyth, Rhys S Allan
The Transcriptional Logic of Mammalian Neuronal Diversity. Ken Sugino, Erin Clark, Anton Schulmann, Yasuyuki Shima, Lihua Wang, David L. Hunt, Bryan M. Hooks, Dimitri Trankner, Jayaram Chandrashekar, Serge Picard, Andrew Lemire, Nelson Spruston, Adam Hantman, Sacha B. Nelson
| Stem cells, regeneration & disease modelling
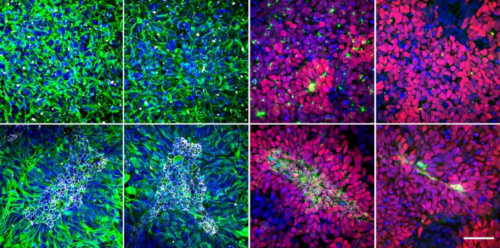
Signaling-dependent control of apical membrane size and self-renewal in rosette-stage human neuroepithelial stem cells. Jan-Philip Medelnik, Kathleen Roensch, Satoshi Okawa, Antonio del Sol, Osvaldo Chara, Levan Mchedlishvili, Elly Tanaka
Fractone bulbs derive from ependymal cells and their laminin composition influence the stem cell niche in the subventricular zone. Marcos Assis Nascimento, Lydia Sorokin, Tatiana Coelho-Sampaio
Comparison of reprogramming factor targets reveals both species-specific and conserved mechanisms in early iPS cells. Kai Fu, Constantinos Chronis, Abdenour Soufi, Giancarlo Bonora, Miguel Edwards, Steve Smale, Kenneth Zaret, Kathrin Plath, Matteo Pellegrini
Differentiation of primate primordial germ cell-like cells following transplantation into the adult gonadal niche. Enrique Sosa, Di Chen, Ernesto J Rojas, Jon Hennebold, Karen Peters, Zhuang Wu, Truong N Lam, Jennifer Mitchell, Ramesh Tailor, Marvin Meistrich, Kyle Orwig, Gunapala Shetty, Amander T Clark
Temporal epigenomic profiling identifies AHR as dynamic super-enhancer controlled regulator of mesenchymal multipotency. Deborah Gerard, Florian Schmidt, Aurelien Ginolhac, Martine Schmitz, Rashi Halder, Peter Ebert, Marcel H. Schulz, Thomas Sauter, Lasse Sinkkonen
Comparison of multiple transcriptomes exposes unified and divergent features of quiescent and activated skeletal muscle stem cells. Natalia Pietrosemoli, Sebastien Mella, Siham Yennek, Meryem Baghdadi, Hiroshi Sakai, Ramkumar Sambasivan, Francesca Pala, DANIELA DI GIROLAMO, Shahragim Tajbakhsh
Single-cell quantitative analysis of skeletal muscle cell population dynamics during regeneration and ageing. Lucia Lisa Petrilli, Filomena Spada, Claudia Fuoco, Elisa Micarelli, Alessio Reggio, Marco Rosina, Cesare Gargioli, Luisa Castagnoli, Gianni Cesareni
Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli are Altered with Age. Mati Mann, Arnav Mehta, Carl de Boer, Monika S. Kowalczyk, Kevin Lee, Noga Rogel, Abigail R. Knecht, Daneyal Farouq, Aviv Regev, David Baltimore
T helper cells modulate intestinal stem cell renewal and differentiation. Moshe Biton, Adam Haber, Semir Beyaz, Noga Rogel, Christopher Smillie, Karthik Shekhar, Alexandra Schnell, Zuojia Chen, Chuan Wu, Jose Ordovas-Montanes, David Alvarez, Rebecca H. Herbst, Itay Tirosh, Grace Burgin, Danielle Dionne, Michael E. Xifaras, Mei Zhang, Alex K. Shalek, Ulrich H. von Andrian, Daniel B. Graham, Orit Rozenblatt-Rosen, Hai Ning Shi, Vijay Kuchroo, Omer Yilmaz, Aviv Regev, Ramnik J. Xavier
The CD11a and EPCR marker combination simplifies and improves the purification of mouse hematopoietic stem cells. Alborz Karimzadeh, Vanessa Scarfone, Connie Chao, Karin Grathwohl, John W. Fathman, David Fruman, Thomas Serwold, Matthew Inla
Instructive starPEG-Heparin biohybrid 3D cultures for modeling human neural stem cell plasticity, neurogenesis, and neurodegeneration. Christos Papadimitriou, Mehmet I Cosacak, Violeta Mashkaryan, Hilal Celikkaya, Laura Bray, Prabesh Bhattarai, Heike Hollak, Xin Chen, Shuijin He, Christopher L Antos, Alvin Thomas, Jens Friedrichs, Andreas Dahl, Yixin Zhang, Uwe Freudenberg, Carsten Werner, Caghan Kizil
Intestinal crypts recover rapidly from focal damage with coordinated motion of stem cells that is impaired by aging. Jiahn Choi, Nikolai Rakhilin, Poornima Gadamsetty, Daniel J. Joe, Tahmineh Tabrizian, Steven M. Lipkin, Derek Huffman, Xiling Shen, Nozomi Nishimura
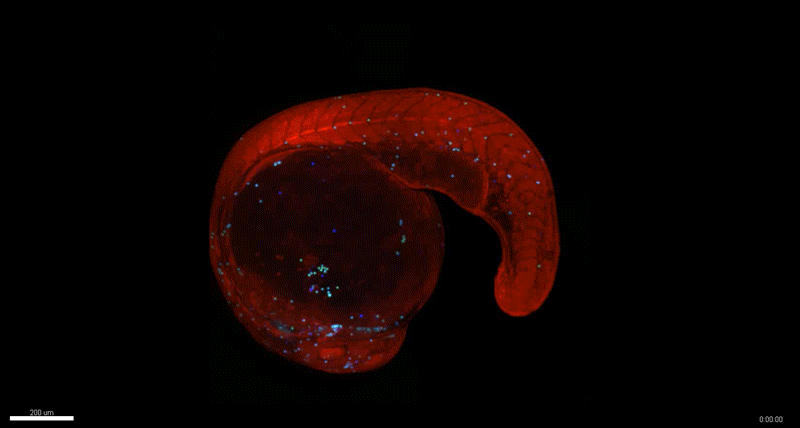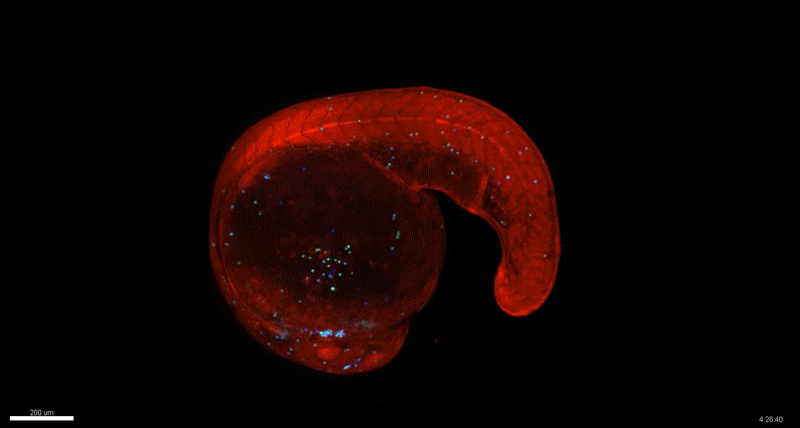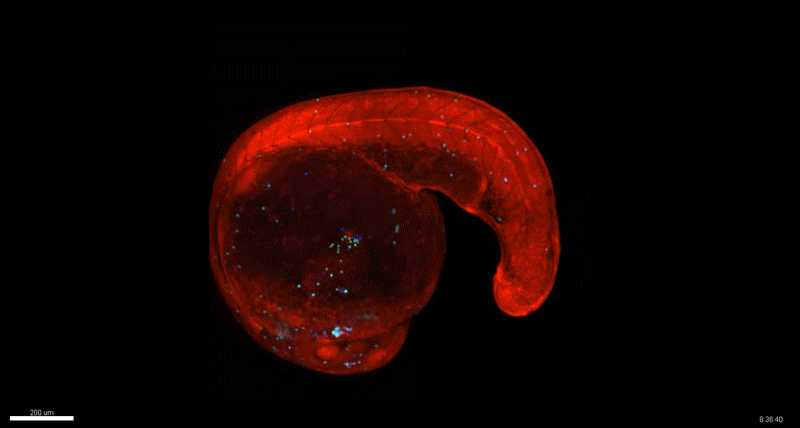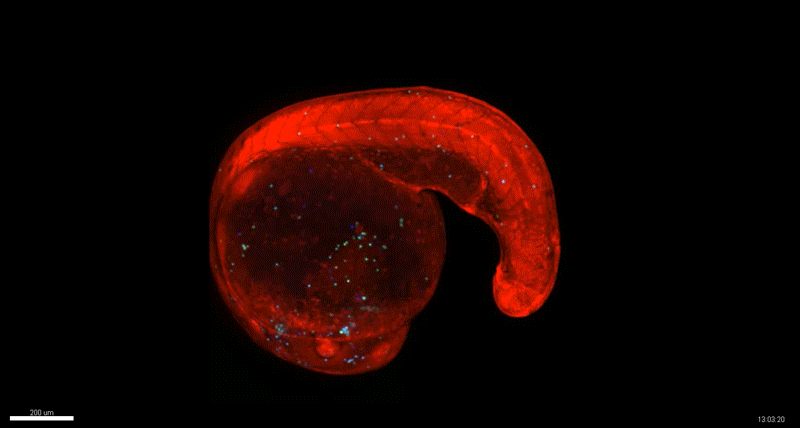
Generation of Mouse-Zebrafish Hematopoietic Tissue Chimeric Embryos for Hematopoiesis and Host-Pathogen Interaction Studies. Margarita M. Parada-Kusz, Anne Clatworthy, Elliott J. Hagedorn, Cristina Penaranda, Anil V. Nair, Jonathan E. Henninger, Christoph Ernst, Brian Li, Raquel Riquelme, Humberto Jijon, Eduardo J. Villablanca, Leonard I. Zon, Deborah Hung, Miguel Allende
Continuous high-resolution in vivo imaging reveals tumor-specific dissemination in an embryonic zebrafish xenograft model. Nandini Asokan, Stephan Daetwyler, Stefanie Nadine Bernas, Christopher Schmied, Steffen Volger, Katrin Lambert, Manja Wobus, Martin Wermke, Gerd Kempermann, Jan Huisken, Michael Brand, Martin Bornhaeuser
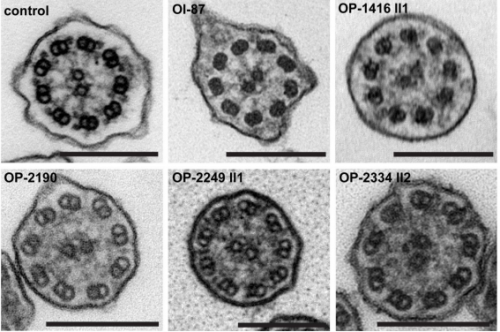
Mutations in C11ORF70 cause primary ciliary dyskinesia with randomization of left/right body asymmetry due to outer and inner dynein arm defects. Inga M Höben, Rim Hjeij, Heike Olbrich, Gerard W Dougherty, Tabea Menchen, Isabella Aprea, Diana Frank, Petra Pennekamp, Bernd Dworniczak, Julia Wallmeier, Johanna Raidt, Kim G Nielsen, Maria C Philipsen, Francesca Santamaria, Laura Venditto, Israel Amirav, Freerk Prenzel, Kaman Wu, Miriam Schmidts, Niki T Loges, Heymut Omran
A phase separated organelle at the root of motile ciliopathy. Ryan Huizar, Chanjae Lee, Alexander Boulgakov, Amjad Horani, Fan Tu, Kevin Drew, Edward Marcotte, Steven Brody, John Wallingford
Phenotyping ciliary dynamics and coordination in response to CFTR-modulators in Cystic Fibrosis respiratory epithelial cells. Maurizio Chioccioli, Luigi Feriani, Jurij Kotar, Preston E. Bratcher, Pietro Cicuta
Gene expression in cord blood links genetic risk for neurodevelopmental disorders with maternal psychological distress and adverse childhood outcomes. Michael S. Breen, Aliza P. Wingo, Nastassja Koen, Kirsten A. Donald, Heather J. Zar, Kerry J. Ressler, Joseph D. Buxbaum, Dan J. Stein
Developmental And Genetic Regulation Of The Human Cortex Transcriptome In Schizophrenia. Andrew E Jaffe, Richard E Straub, Joo Heon Shin, Ran Tao, Yuan Gao, Leonardo Collado Torres, Tony Kam-Thong, Hualin S Xi, Jie Quan, Qiang Chen, Carlo Colantuoni, William S Ulrich, Brady J Maher, Amy Deep-Soboslay, The BrainSeq Consortium, Alan Cross, Nicholas J Brandon, Jeffrey T Leek, Thomas M Hyde, Joel E Kleinman, Daniel R Weinberger
Gene expression imputation across multiple brain regions reveals schizophrenia risk throughout development. Laura M Huckins, Amanda Dobbyn, Douglas Ruderfer, Gabriel Hoffman, Weiqing Wang, Antonio F Pardinas, Veera M Rajagopal, Thomas D Als, Hoang Tan Hoang, Kiran Girdhar, James Boocock, Panagiotis Roussos, Menachem Fromer, Robin Kramer, Enrico Domenici, Eric Gamazon, Shaun Purcell, CommonMind Consortium, Schizophrenia Working Group of the Psychiatric Gen, iPSYCH-GEMS Schizophrenia Working Group, Ditte Demontis, Anders Borglum, James Walters, Michael O’Donovan, Patrick F Sullivan, Micahel Owen, Bernie Devlin, Solveig K Sieberts, Nancy Cox, Hae Kyung Im, Pamela Sklar, Eli Ayumi Stahl
Germ line aging and regional epigenetic instability: age prediction using human sperm DNA methylation signatures. Timothy G Jenkins, Kenneth I Aston, Douglas T Carrell
Aging and neurodegeneration are associated with increased mutations in single human neurons. Michael A Lodato, Rachel E Rodin, Craig L Bohrson, Michael E Coulter, Alison R Barton, Minseok Kwon, Maxwell A Sherman, Carl M Vitzhum, Lovelace J Luquette, Chandri Yandava, Pengwei Yang, Thomas W Chittenden, Nicole E Hatem, Steven C Ryu, Mollie B Woodworth, Peter J Park, Christopher A Walsh
Epigenetic influences on aging: a longitudinal genome-wide methylation study in old Swedish twins. Yunzhang Wang, Robert Karlsson, Erik Lampa, Qian Zhang, Åsa K. Hedman, Malin Almgren, Catarina Almqvist, Allan F. McRae, Riccardo Marioni, Erik Ingelsson, Peter M. Visscher, Ian J. Deary, Lars Lind, Tiffany Morris, Stephan Beck, Nancy L. Pedersen, Sara Hägg
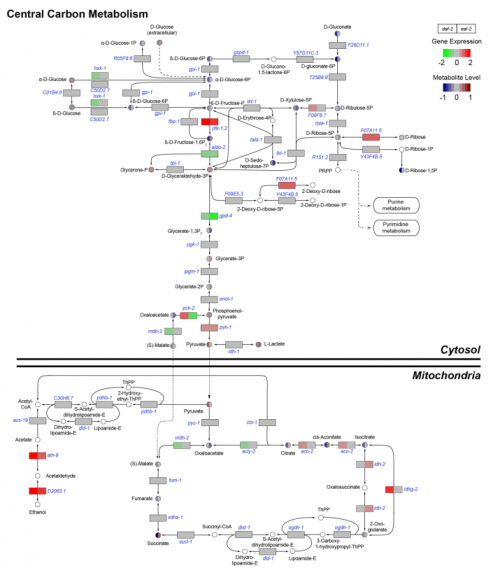
Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Arwen Gao, Reuben Smith, Michel Van Weeghel, Rashmi Kamble, Riekelt Houtkooper
An essential post-developmental role for Lis1 in mice. Timothy J Hines, Xu Gao, Subhshri Sahu, Meghann M Lange, Jill R Turner, Jeffery L Twiss, Deanna S Smith
A urine-dependent human urothelial organoid offers a promising alternative to rodent models of infection. Harry Horsley, Dhanuson Dharmasena, James Malone-Lee, Jennifer L. Rohn
Germline loss of MBD4 predisposes to leukaemia due to a mutagenic cascade driven by 5mC. Mathijs A. Sanders, Edward Chew, Christoffer Flensburg, Annelieke Zeilemaker, Sarah E. Miller, Adil al Hinai, Ashish Bajel, Bram Luiken, Melissa Rijken, Tamara Mclennan, Remco M. Hoogenboezem, François G. Kavelaars, Marnie E. Blewitt, Eric M. Bindels, Warren S. Alexander, Bob Löwenberg, Andrew W. Roberts, Peter J. M. Valk, Ian Majewski
An Essential Role for Argonaute 2 in EGFR-KRAS Signaling in Pancreatic Cancer Development. Sunita Shankar, Jean Tien, Ronald F Siebenaler, Vijaya L Dommeti, Sylvia Zelenka-Wang, Kristin M Juckette, Alice Xu, Malay Mody, Andrew Goodrum, Grace Tsaloff, Ingrid J Apel, Lisha Wang, Javed Siddiqui, Jiaqi Shi, Chandan Kumar-Sinha, Arul M Chinnaiyan
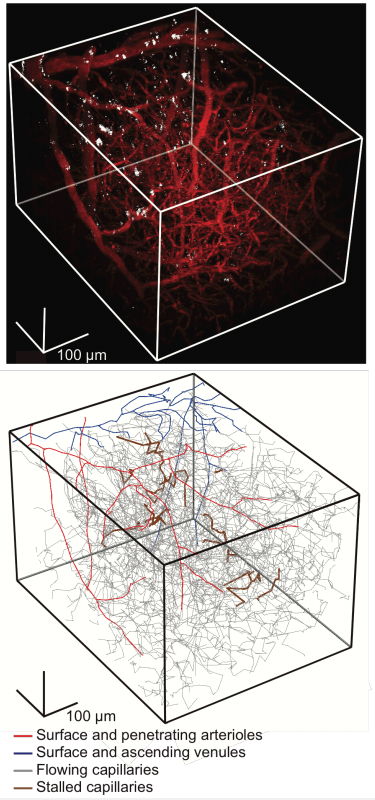
Neutrophil adhesion in brain capillaries contributes to cortical blood flow decreases and impaired memory function in a mouse model of Alzheimer’s disease. Jean C. Cruz Hernandez, Oliver Bracko, Calvin J. Kersbergen, Victorine Muse, Mohammad Haft-Javaherian, Maxime Berg, Laibaik Park, Lindsay K. Vinarcsik, Iryna Ivasyk, Yiming Kang, Marta Cortes-Canteli, Myriam Peyrounette, Vincent Doyeux, Amy Smith, Joan Zhou, Gabriel Otte, Jeffrey D. Beverly, Elizabeth Davenport, Yohan Davit, Sidney Strickland, Costantino Iadecola, Sylvie Lorthois, Nozomi Nishimura, Chris B. Schaffer
PPARD regulation in gastric progenitor cells drives gastric tumorigenesis in mice. Xiangsheng Zuo, Yasunori Deguchi, Weiguo Xu, Daoyan Wei, Rui Tian, Weidong Chen, Micheline J. Moussalli, Yi Liu, Fei Mao, Min Xu, Yaying Yang, Shen Gao, Jonathan C. Jaoude, Fuyao Liu, Mihai Gagea, Russell Broaddus, Keping Xie, Imad Shureiqi
L-tyrosine supplementation is not therapeutic for skeletal muscle dysfunction in zebrafish and mouse models of dominant skeletal muscle α-actin nemaline myopathy. Adriana M. Messineo, Charlotte Gineste, Tamar E. Sztal, Elyshia L McNamara, Christophe Vilmen, Augustin C. Ogier, Dorothee Hahne, David Bendahan, Nigel G. Laing, Robert J. Bryson-Richardson, Julien Gondin, Kristen J. Nowak
Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes. David A. Knowles, Courtney K. Burrows, John D. Blischak, Kristen M. Patterson, Carole Ober, Jonathan K. Pritchard, Yoav Gilad
Endothelial Cell-Specific MCPIP Deletion Causes Endothelial Dysfunction and Impairs Post-ischemic Angiogenesis in Vivo. Jianli Niu, Nidhi Kapoor, Jian Liang, Zhuqing Jin, Edilu Becerra, Pappachan E. Kolattukudy
PKD2 is an essential ion channel subunit in the primary cilium of the renal collecting duct epithelium. Xiaowen Liu, Thuy Vien, Jingjing Duan, Shu-Hsien Sheu, Paul G. DeCaen, David E. Clapham
Cadmium exposure inhibits branching morphogenesis and causes alterations consistent with HIF-1α inhibition in human primary breast organoids. Sabrina A. Rocco, Lada Koneva, Lauren Y. M. Middleton, Tasha Thong, Sumeet Solanki, Sarah Karram, Kowit Nambunmee, Craig Harris, Laura S. Rozek, Maureen A. Sartor, Yatrik M. Shah, Justin Colacino
Genomic Locus Modulating Corneal Thickness in the Mouse Identifies POU6F2 as a Potential Risk of Developing Glaucoma. Rebecca King, Felix L. Struebing, Ying Li, Jiaxing Wang, Allison Ashley Koch, Jessica Cooke Bailey, Puya Gharahkhan, Stuart MacGregor, R. Rand Allingham, Michael A. Hauser, Janey L. Wiggs, Eldon E. Geisert
Clonal analysis of newborn hippocampal dentate granule cell proliferation and development in temporal lobe epilepsy. Shatrunjai P. Singh, Candi L. LaSarge, Amen An, John J. McAuliffe, Steve C. Danzer
Evo-devo & evo
Hox genes pattern the primary body axis of an anthozoan cnidarian prior to gastrulation. Timothy Q. DuBuc, Thomas B. Stephenson, Amber N. Rock, Mark Q. Martindale
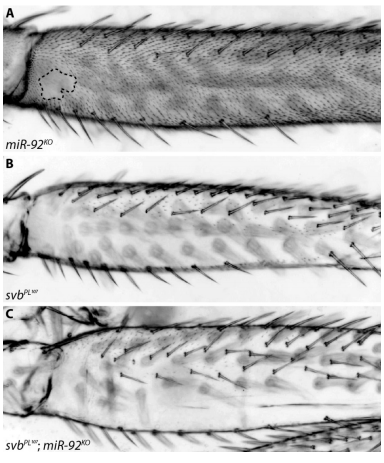
Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. Sebastian Kittelmann, Alexandra D. Buffry, Isabel Almudi, Marianne Yoth, Gonzalo Sabaris, Franziska A. Franke, Juan Pablo Couso, Maria D. S. Nunes, Nicolas Frankel, Jose Luis Gomez-Skarmeta, Jose Pueyo-Marques, Saad Arif, Alistair P. McGregor
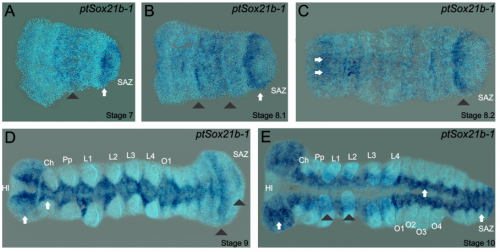
Duplication and divergence of Sox genes in spiders. Christian L. B. Paese, Daniel J. Leite, Anna Schoenauer, Alistair P. McGregor, Steven Russell
Pervasive correlated evolution in gene expression shapes cell type transcriptomes. Cong Liang, Jacob M. Musser, Alison Cloutier, Richard Prum, Gunter Wagner
How morphological development can guide evolution. Sam Kriegman, Nick Cheney, Josh Bongard
Developmental loci harbor clusters of accelerated regions that evolved independently in ape lineages. Dennis Kostka, Alisha K. Holloway, Katherine Pollard
Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Marta Florio, Michael Heide, Holger Brandl, Anneline Pinson, Sylke Winkler, Pauline Wimberger, Wieland B. Huttner, Michael Hiller
Developmental constraints on genome evolution in four bilaterian model species. Jialin Liu, Marc Robinson-Rechavi
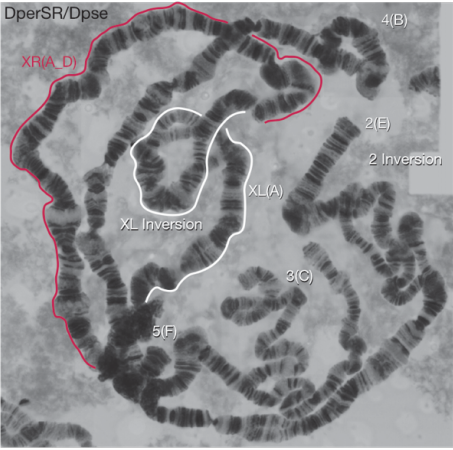
The role of chromosomal inversions in speciation. Zachary Fuller, Christopher Leonard, Randee Young, Stephen Schaeffer, Nitin Phadnis
Gene regulatory network rewiring by an adaptively evolving microRNA cluster in Drosophila. Yang Lyu, Zhongqi Liufu, Juan Xiao, Yuxin Chen, Chung-I Wu, Tian Tang
Rapid evolution of gained essential developmental functions of a young gene via interactions with other essential genes. Yuh Chwen G Lee, Iuri M Ventura, Gavin R Rice, Don-Yuan Chen, Manyuan Long
The asexual genome of Drosophila. Stephan Schiffels, Ville Mustonen, Michael Lässig
Improved genome assembly and annotation for the rock pigeon (Columba livia). Carson Holt, Michael Campbell, David Keays, Nathaniel Edelman, Aurelie Kapusta, Emily Maclary, Eric Domyan, Alexander Suh, Wesley Warrenb, Mark Yandell, Thomas Gilbert, Michael D. Shapiro
Hemimetabolous genomes reveal molecular basis of termite eusociality. Mark C. Harrison, Evelien Jongepier, Hugh M. Robertson, Nicolas Arning, Tristan Bitard-Feildel, Hsu Chao, Christopher P. Childers, Huyen Dinh, Harshavardhan Doddapaneni, Shannon Dugan, Johannes Gowin, Carolin Greiner, Yi Han, Haofu Hu, Daniel S. T. Hughes, Ann-Kathrin Huylmans, Carsten Kemena, Lukas P. M. Kremer, Sandra L. Lee, Alberto Lopez-Ezquerra, Ludovic Mallet, Jose M. Monroy-Kuhn, Annabell Moser, Shwetha C. Murali, Donna M. Muzny, Saria Otani, Maria-Dolors Piulachs, Monica Poelchau, Jiaxin Qu, Florentine Schaub, Ayako Wada-Katsumata, Kim C. Worley, Qiaolin Xie, Guillem Ylla, Michael Poulsen, Richard A. Gibbs, Coby Schal, Stephen Richards, Xavier Belles, Judith Korb, Erich Bornberg-Bauer
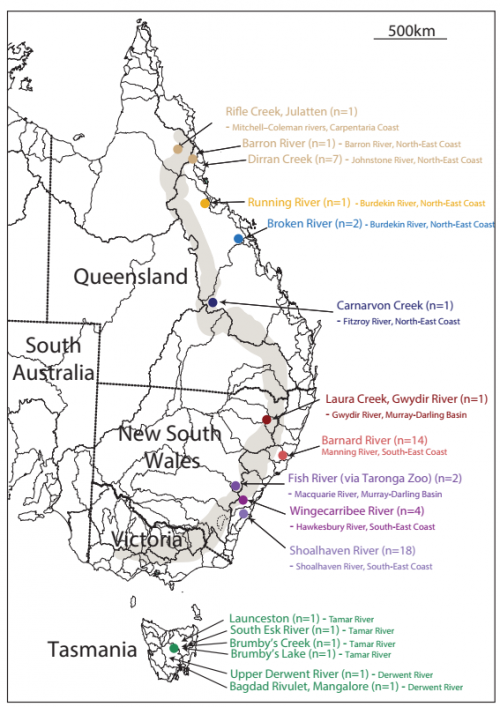
Insights into platypus population structure and history from whole-genome sequencing. Hilary C Martin, Elizabeth M Batty, Julie Hussin, Portia Westall, Tasman Daish, Stephen Kolomyjec, Paolo Piazza, Rory Bowden, Margaret Hawkins, Tom Grant, Craig Moritz, Frank Grutzner, Jaime Gongora, Peter Donnelly
Reduction in major transcription factor binding domain families during the evolution of the avian lineage. Allie M. Graham, Jason S. Presnell
Host shifts result in parallel genetic changes when viruses adapt to closely related species. Ben Longdon, Jonathan P Day, Joel M Alves, Sophia CL Smith, Thomas M Houslay, John E McGonigle, Lucia Tagliaferri, Francis M Jiggins
Overlooked polyploidies in lycophytes generalize their roles during the evolution of vascular plants. Jinpeng Wang, Jigao Yu, Pengchuan Sun, Chao Li, Xiaoming Song, Tianyu Lei, Yuxian Li, Jiaqing Yuan, Sangrong Sun, Hongling Ding, Xueqian Duan, Shaoqi Shen, Yanshuang Shen, Jing Li, Fanbo Meng, Yangqin Xie, Jianyu Wang, Yue Hou, Jin Zhang, Xianchun Zhang, Xiyin Wang
A primitive starfish ancestor from the Early Ordovician of Morocco reveals the origin of crown group Echinodermata. Aaron W. Hunter, Javier Ortega-Hernandez
Russian Doll Genes and Complex Chromosome Rearrangements in Oxytricha trifallax. Jasper Braun, Lukas Nabergall, Rafik Neme, Laura F Landweber, Masahico Saito, Natasha Jonoska
Nearly Neutral Evolution Across the Drosophila melanogaster Genome. David Castellano, Jennifer James, Adam Eyre-Walker
Cell biology
Formation and disassembly of a contractile actomyosin network mediates content release from large secretory vesicles. Dagan Segal, Assaf Zaritsky, Eyal D. Schejter, Ben Zion Shilo
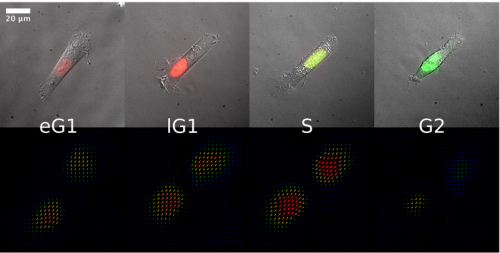
Variation in traction forces during cell cycle progression. Benoit Vianay, Fabrice Senger, Simon Alamos, Maya Anjur-Dietrich, Elizabeth Bearce, Bevan Cheeseman, Lisa Lee, Manuel Thery
Cell size control driven by the circadian clock and environment in cyanobacteria. Bruno M. C. Martins, Amy K. Tooke, Philipp Thomas, James C. W. Locke
RodZ promotes MreB polymer formation and curvature localization to determine the cylindrical uniformity of E. coli shape. Randy M Morgenstein, Benjamin P Bratton, Joshua W Shaevitz, Zemer Gitai
Giant flagellins form thick flagellar filaments in two species of marine Gammaproteobacteria. Nicholas Maitland Thomson, Josie L Ferreira, Teige R Matthews-Palmer, Morgan Beeby, Mark J Pallen
Uncoupling Intraflagellar Transport and Primary Cilia Formation Demonstrates Deep Integration of IFT in Hedgehog Signaling. Thibaut Eguether, Fabrice P Cordelieres, Gregory J Pazour
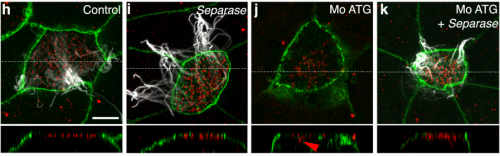
CDC20B is required for deuterosome-mediated centriole production in multiciliated cells. Diego R. Revinski, Laure-Emmanuelle Zaragosi, Camille Boutin, Sandra Ruiz-Garcia, Marie Deprez, Olivier Rosnet, Virginie Thome, Olivier Mercey, Agnes Paquet, Nicolas Pons, Brice Marcet, Laurent Kodjabachian, Pascal Barbry
Structural centrosome aberrations promote non-cell-autonomous invasiveness. Olivier Ganier, Dominik Schnerch, Philipp Oertle, Roderick Y. H. Lim, Marija Plodinec, Erich A. Nigg
A robust and tunable mitotic oscillator in artificial cells. Ye Guan, Zhengda Li, Shiyuan Wang, Patrick M. Barnes, Xuwen Liu, Haotian Xu, Minjun Jin, Allen P. Liu, Qiong Yang
Exportin Crm1 is repurposed as a docking protein to generate microtubule organizing centers at the nuclear pore. Xun Bao, Christos Spanos, Tomoko Kojidani, Eric Lynch, Juri Rappsilber, Yasushi Hiraoka, Tokuko Haraguchi, Kenneth Sawin
Endocytosis caused by liquid-liquid phase separation of proteins. Louis-Philippe Bergeron-Sandoval, Hossein Khadivi Heris, Adam G. Hendricks, Allen J. Ehrlicher, Paul Francois, Rohit V. Pappu, Stephen W. Michnick
Systematic analysis of the molecular architecture of endocytosis reveals a nanoscale actin nucleation template that drives efficient vesicle formation. Markus Mund, Johannes Albertus van der Beek, Joran Deschamps, Serge Dmitrieff, Jooske Louise Monster, Andrea Picco, Francois Nedelec, Marko Kaksonen, Jonas Ries
The contributions of the actin machinery to endocytic membrane bending and vesicle formation. Andrea Picco, Wanda Kukulski, Hetty E. Manenschijn, Tanja Specht, John A. G. Briggs, Marko Kaksonen

CYRI (FAM49) proteins are local inhibitors of Scar/WAVE induced lamellipodia that bind directly to active Rac1. Loic Fort, Jose Batista, Peter A Thomason, Heather J Spence, Jennifer Greaves, Kirsty J Martin, Kurt I Anderson, Peter Brown, Sergio Lilla, Matthwe P Neilson, Petra Tafelmeyer, Sara Zanivan, Shehab Ismail, Nicholas C.O. Tomkinson, Luke H Chamberlain, Robert H Insall, Laura M Machesky
Dynein dynamics at the microtubule plus-ends and cortex during division in the C. elegans zygote. Ruddi Rodriguez-Garcia, Laurent Chesneau, Sylvain Pastezeur, Julien Roul, Marc Tramier, Jacques Pecreaux
The tumor suppressor APC is an attenuator of spindle-pulling forces during C. elegans asymmetric cell division. Kenji Sugioka, Lars-Eric Fielmich, Kota Mizumoto, Bruce Bowerman, Sander van den Heuvel, Akatsuki Kimura, Hitoshi Sawa
Dissecting single cell molecular spatiotemporal mobility and clustering at Focal Adhesions in polarised cells by fluorescence fluctuation spectroscopy methods. Jorge Bernardino de la Serna, Esther Garcia
Stopping Transformed Growth with Cytoskeletal Proteins: Turning a Devil into an Angel. Bo Yang, Haguy Wolfenson, Naotaka Nakazawa, Shuaimin Liu, Junqiang Hu, Michael Sheetz
Modelling
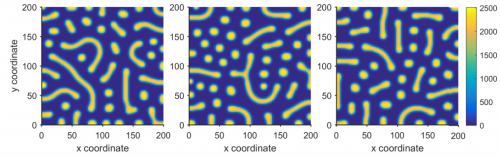
Model reduction permits Turing instability analysis of arbitrary reaction-diffusion models. Stephen Smith, Neil Dalchau
The phenomenon of growing surface interference explains the rosette pattern of jaguar. Michäel Dougoud, Christian Mazza, Beat Schwaller, Laszlo Pecze
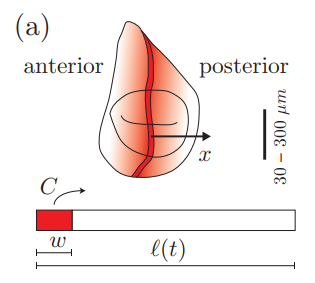
Dynamic gradient scaling as a critical point of growth control. Daniel Aguilar Hidalgo, Steffen Werner, Ortrud Wartlick, Marcos González-Gaitán, Benjamin Friedrich, Frank Jülicher
Temporal precision of regulated gene expression. Shivam Gupta, Julien Varennes, Hendrik C. Korswagen, Andrew Mugler
A biophysical model explains the spontaneous bursting behavior in the developing retina. Dora Karvouniari, Lionel Gil, Olivier Marre, Serge Picaud, Bruno Cessac
An efficient gene regulatory network inference algorithm for early Drosophila melanogaster embryogenesis. Hirotaka Matsumoto, Hisanori Kiryu, Yasuhiro Kojima, Suguru Yaginuma, Itoshi Nikaido
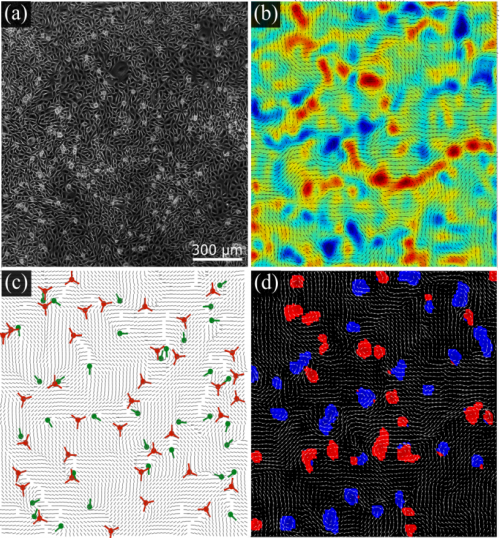
Turbulent dynamics of epithelial cell cultures. C. Blanch-Mercader, V. Yashunsky, S. Garcia, G. Duclos, L. Giomi, P. Silberzan
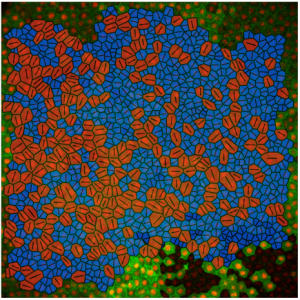
Mechanical characterisation of disordered and anisotropic cellular monolayers. Alexander Nestor-Bergmann, Emma Johns, Sarah Woolner, Oliver E. Jensen
Do cells sense time by number of divisions? Zeev Schuss, Kimsy Tor, David Holcman
Euplotid: A quantized geometric model of the eukaryotic cell. Diego Borges-Rivera
Noise-driven Cell Differentiation and the Emergence of Organization. Hadiseh Safdari, Ata Kalirad, Cristian Picioreanu, Rouzbeh Tusserkani, Mehdi Sadeghi
A biophysical dynamic module for the polarisation of auxin efflux carriers PIN-FORMED (PIN). Valeria Hernandez-Hernandez, Rafael A. Barrio, Mariana Benitez, Naomi Nakayama, Jose Roberto Romero-Arias, Carlos Villarreal
Discrete and continuum approximations for collective cell migration in a scratch assay with cell size dynamics. Oleksii M Matsiaka, Catherine Penington, Ruth E Baker, Matthew Simpson
Shaping Epigenetic Memory via Genomic Bookmarking. Davide Michieletto, Michael Chiang, Davide Coli, Argyris Papantonis, Enzo Orlandini, Peter R. Cook, Davide Marenduzzo
Subtle alteration in microRNA dynamics accounts for differential nature of cellular proliferation. Dola Sengupta, Vinodhini Govindaraj, Sandip Kar
Mathematics at the eve of a historic transition in biology. Guo-Wei Wei
Tools & resources
| Imaging etc.
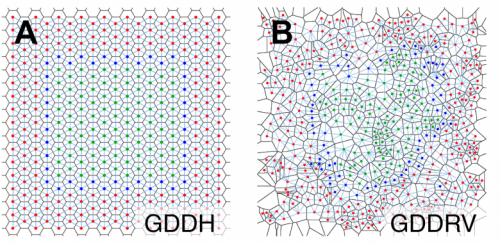
EpiGraph: an open-source platform to quantify epithelial organization. Pablo Vicente-Munuera, Pedro Gomez-Galvez, Antonio Tagua, Marta Letran, Yanlan Mao, Luis M. Escudero
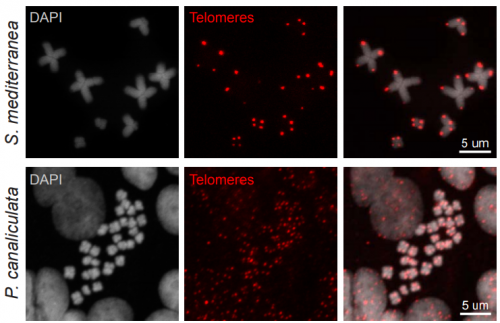
An adaptable chromosome preparation methodology for use in invertebrate research organisms. Longhua Guo, Alice Accorsi, Shuonan He, Carlos Guerrero-Hernández, Shamilene Sivagnanam, Sean McKinney, Matthew Gibson, Alejandro Sánchez Alvarado
Combined flow cytometry and high throughput image analysis for the study of essential genes in Caenorhabditis elegans. Blanca Hernando-Rodríguez, Annmary Paul Erinjeri, María Jesús Rodríguez-Palero, Val Millar, Sara González-Hernández, María Olmedo, Bettina Schulze, Ralf Baumeister, Manuel J. Muñoz, Peter Askjaer, Marta Artal-Sanz
A method to keep the excitation light sheet in focus in selective plane illumination microscopy. Liang Gao
Topography and refractometry of sperm cells using SLIM. Lina Liu, Mikhail E Kandel, Marcello Rubessa, Sierra Schreiber, Mathew Wheeler, Gabriel Popescu
Simultaneously measuring image features and resolution in live-cell STED images. Andrew E. S. Barentine, Lena K. Schroeder, Michael Graff, David Baddeley, Joerg Bewersdorf
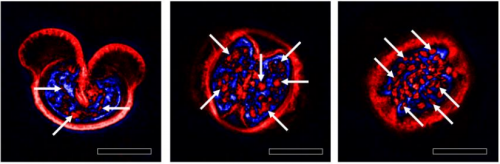
3D label-free imaging and analysis of Pinus pollen grains using optical diffraction tomography. Geon Kim, SangYun Lee, Seungwoo Shin, YongKeun Park
Three dimensional cross-modal image inference: label-free methods for subcellular structure prediction. Chek Ounkomol, Daniel A. Fernandes, Sharmishtaa Seshamani, Mary M. Maleckar, Forrest Collman, Gregory R. Johnson
Optical crosstalk in SPAD arrays for high-throughput single-molecule fluorescence spectroscopy. Antonino Ingargiola, Maya Segal, Angelo Gulinatti, Ivan Rech, Ivan Labanca, Piera Maccagnani, Massimo Ghioni, Shimon Weiss, Xavier Michalet
Recording neural activity in unrestrained animals with 3D tracking two photon microscopy. Doycho Karagyozov, Mirna Mihovilovic Skanata, Amanda Lesar, Marc Gershow
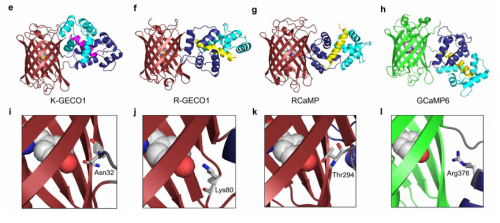
A genetically encoded Ca2+ indicator based on circularly permutated sea anemone red fluorescent protein. Yi Shen, Hod Dana, Ahmed S. Abdelfattah, Ronak Patel, Jamien Shea, Rosana S. Molina, Bijal Rawal, Vladimir Rancic, Yu-Fen Chang, Lanshi Wu, Yingche Chen, Yong Qian, Matthew D. Wiens, Nathan Hambleton, Klaus Ballanyi, Thomas E. Hughes, Mikhail Drobizhev, Douglas S. Kim, Minoru Koyama, Eric R. Schreiter, Robert E. Campbell
A FRET based biosensor for measuring Gα13 activation in single cells. Marieke Mastop, Nathalie R. Reinhard, Cristiane R. Zuconelli, Fenna Terwey, Theodorus W. J. Gadella Sr., Jakobus van Unen, Merel J. W. Adjobo-Hermans, Joachim Goedhart
Understanding boundary effects and confocal optics enables quantitative FRAP analysis in the confined geometries of animal, plant, and fungal cells. James L Kingsley, Jeffrey P Bibeau, Iman Mousavi, Cem Unsal, Zhilu Chen, Xinming Huang, Luis Vidali, Erkan Tuzel
Multidimensional quantitative analysis of mRNA expression within intact vertebrate embryos. Vikas Trivedi, Harry M. T. Choi, Scott E. Fraser, Niles A. Pierce
Single-molecule fluorescent amplification of RNA using clampFISH probes. Sara H Rouhanifard, Margaret Dunagin, Ian A Mellis, Sareh Bayatpour, Orsolya Symmons, Allison Cote, Arjun Raj
An optimized pipeline for parallel image-based quantification of gene expression and genotyping after in situ hybridization. Tomasz Dobrzycki, Monika Krecsmarik, Florian Bonkhofer, Roger K Patient, Rui Monteiro
Quantitative methods to investigate the 4D dynamics of heterochromatic repair sites in Drosophila cells. Christopher P Caridi, Laetitia Delabaere, Harianto Tjong, Hannah Hopp, Devika Das, Frank Alber, Irene Chiolo
In vivo multiphoton fluorescence imaging with polymer dots. Ahmed M Hassan, Xu Wu, Jeremy W Jarrett, Shihan Xu, David R Miller, Jiangbo Yu, Evan P Perillo, Yen-Liang Liu, Daniel T Chiu, Hsin-Chih Yeh, Andrew K Dunn

Eukaryotically expressed encapsulins as orthogonal compartments for multiscale molecular imaging. Felix Sigmund, Christoph Massner, Philipp Erdmann, Anja Stelzl, Hannes Rolbieski, Helmut Fuchs, Martin Hrabe de Angelis, Mitul Desai, Sarah Bricault, Alan Jasanoff, Vasilis Ntziachristos, Juergen Plitzko, Gil Gregor Westmeyer
Brillouin microscopy, what is it really measuring? Pei-Jung Wu, Irina Kabakova, Jeffrey Ruberti, Joseph M. Sherwood, Iain E. Dunlop, Carl Paterson, Peter Török, Darryl R. Overby
REASSESSING THE REVOLUTIONS RESOLUTIONS. Marin van Heel, Michael Schatz
Simultaneous Determination of Protein Structure and Dynamics Using Cryo-Electron Microscopy. Massimiliano Bonomi, Riccardo Pellarin, Michele Vendruscolo
ShuTu: Open-Source Software for Efficient and Accurate Reconstruction of Dendritic Morphology. Dezhe Z. Jin, Ting Zhao, David Hunt, Rachel Pearcy, Ching-Lung Hsu, Nelson Spruston
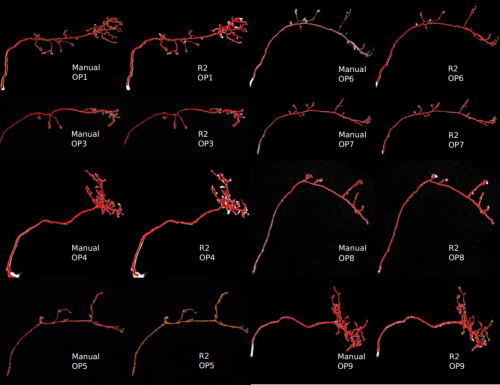
Automated 3D Neuron Tracing with Precise Branch Erasing and Confidence Controlled Back-Tracking. Siqi Liu, Donghao Zhang, Yang Song, Hanchuan Peng, Weidong Cai
MICCS: A Fully Programmable Multipurpose Integrated Cell Culture System. Timothy Kassis, Paola M. Perez, Chloe J. W. Yang, Luis R. Soenksen, David L. Trumper, Linda G. Griffith
Visual Representation of Experimental Protocols. James Scott-Brown, Antonis Papachristodoulou
Protein structures as shapes: Analysing protein structure variation using geometric morphometrics. Jose Sergio Hleap, Christian Blouin
Which line to follow? The utility of different line-fitting methods to capture the mechanism of morphological scaling. Alexander W. Shingleton
A Cautionary Tail: Changes in Integrin Behavior with Labeling. Catherine G. Galbraith, Michael W. Davidson, James A. Galbraith
Exploring biological networks in 3D, stereoscopic 3D and immersive 3D with iCAVE. Selim Kalayci, Zeynep H. Gumus
Quantitative imaging of chromatin decompaction in living cells. Elisa Dultz, Roberta Mancini, Guido Polles, Pascal Vallotton, Frank Alber, Karsten Weis
| Genome tools
CRISPys: Optimal sgRNA design for editing multiple members of a gene family using the CRISPR system. Gal Hyams, Shiran Abadi, Adi Avni, Eran Halperin, Eilon Shani, Itay Mayrose
Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Qiang Zhang, Hui-Li Xing, Zhi-Ping Wang, Hai-Yan Zhang, Fang Yang, Xue-Chen Wang, Qi-Jun Chen
CRISPR-Cas12a target binding unleashes single-stranded DNase activity. Janice S Chen, Enbo Ma, Lucas B Harrington, Xinran Tian, Jennifer A Doudna
SgTiler: a fast method to design tiling sgRNAs for CRISPR/Cas9 mediated screening. Musaddeque Ahmed, Housheng Hansen He
Easy quantification of template-directed CRISPR/Cas9 editing. Eva Karina Brinkman, Arne Nedergaard Kousholt, Tim Harmsen, Christ Leemans, Tao Chen, Jos Jonkers, Bas van Steensel
Covalent linkage of the DNA repair template to the CRISPR/Cas9 complex enhances homology-directed repair. Natasa Savic, Femke Ringnalda, Katja Bargsten, Yizhou Li, Christian Berk, Jonathan Hall, Dario Neri, Martin Jinek, Gerald Schwank
Rapid, modular, and cost-effective generation of donor DNA constructs for CRISPR-based gene knock-in. Yi-Jiun Chen, Weikang Wang, Xiao-Jiun Tian, Daniel E. Lefever, David A. Taft, Jianhua Xing
Transgenic mouse lines expressing the 3xFLAG-dCas9 protein for enChIP analysis. Toshitsugu Fujita, Fusako Kitaura, Asami Oji, Naoki Tanigawa, Miyuki Yuno, Masahito Ikawa, Ichiro Taniuchi, Hodaka Fujii
A suite of transgenic driver and reporter mouse lines with enhanced brain cell type targeting and functionality. Tanya L Daigle, Linda Madisen, Travis A Hage, Matthew T Valley, Ulf Knoblich, Rylan S Larsen, Marc M Takeno, Lawrence Huang, Hong Gu, Rachael Larsen, Maya Mills, Alice Bosma-Moody, La’Akea Siverts, Miranda Walker, Lucas T Graybuck, Zizhen Yao, Olivia Fong, Emma Garren, Garreck Lenz, Mariya Chavarha, Julie Pendergraft, James Harrington, Karla E Hirokawa, Julie A Harris, Medea McGraw, Douglas R Ollerenshaw, Kimberly Smith, Baker A Baker, Jonathan T Ting, Susan M Sunkin, Jerome Lecoq, Michael Z Lin, Edward S Boyden, Gabe J Murphy, Nuno da Costa, Jack Waters, Lu Li, Bosiljka Tasic, Hongkui Zeng
A versatile genetic tool for post-translational control of gene expression in Drosophila melanogaster. Sachin Sethi, Jing W. Wang
Saturating Single-Cell Datasets. Aparna Bhaduri, Tomasz J. Nowakowski, Alex A. Pollen, Arnold R. Kriegstein
Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Nelly Olova, Felix Krueger, Simon Andrews, David Oxley, Rebecca V Berrens, Miguel R Branco, Wolf Reik
Rapid high-resolution measurement of DNA replication timing by droplet digital PCR. Dzmitry G. Batrakou, Emma D. Heron, Conrad A. Nieduszynski
Improving Gene Regulatory Network Inference by Incorporating Rates of Transcriptional Change.Jigar Desai, Ryan C. Sartor, Lovely Mae Lawas, Krishna S. V. Jagadish, Colleen J. Doherty
Removing unwanted variation between samples in Hi-C experiments. Kipper Fletez-Brant, Yunjiang Qiu, David U Gorkin, Ming Hu, Kasper D Hansen
Binless normalization of Hi-C data provides significant interaction and difference detection independently of resolution. Yannick G. Spill, David Castillo, Marc A. Marti-Renom
Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Luca Venturini, Shabhonam Caim, Gemy Kaithakottil, Daniel Lee Mapleson, David Swarbreck
Spliceosome profiling visualizes the operations of a dynamic RNP in vivo at nucleotide resolution. Jordan Burke, Adam Longhurst, Daria Merkurjev, Jade Sales-Lee, Beiduo Rao, James Moresco, John Yates III, Jingyi Jessica Li, Hiten D Madhani
Transcriptome-wide analysis of the functional intronome using spliceosome profiling. Weijun Chen, Jill Moore, Hakan Ozadam, Hennady P Shulha, Nicholas Rhind, Zhiping Weng, Melissa J. Moore
TF2Network: predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Shubhada R. Kulkarni, Dries Vaneechoutte, Jan Van de Velde, Klaas Vandepoele
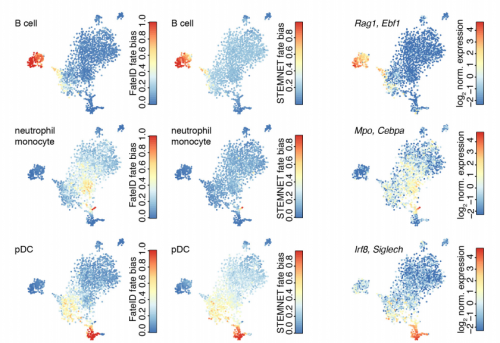
FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Josip Stefan Herman, Sagar Sagar, Dominic Grun
Cross-Site Comparison of Ribosomal Depletion Kits for Illumina RNAseq Library Construction. Zachary T Herbert, Jamie P Kershner, Vincent L Butty, Jyothi Thimmapuram, Sulbha Choudhari, Yuriy O Alekseyev, Jun Fan, Jessica W Podnar, Edward Wilcox, Jenny Gipson, Allison Gillaspy, Kristen Jepsen, Sandra Splinter BonDurant, Krystalynne Morris, Maura Berkeley, Ashley LeClerc, Stephen D. Simpson, Gary Sommerville, Leslie Grimmett, Marie Adams, Stuart S. Levine
An interlaboratory study of complex variant detection. Stephen E Lincoln, Justin M Zook, Shimul Chowdhury, Shazia Mahamdallie, Andrew Fellowes, Eric W Klee, Rebecca Truty, Catherine Huang, Farol L Tomson, Megan H Cleveland, Peter M Vallone, Yan Ding, Sheila Seal, Wasanthi DeSilva, Russell K Garlick, Marc Salit, Nazneen Rahman, Stephen F Kingsmore, Swaroop Aradhya, Robert L Nussbaum, Matthew J Ferber, Brian H Shirts
MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Daniel Huson, Benjamin Albrecht, Caner Bagci, Irina Bessarab, Anna Gorska, Dino Jolic, Rohan B.H. Williams
CREAM: Clustering of genomic REgions Analysis Method. Seyed Ali Madani Tonekaboni, Parisa Mazrooei, Victor Kofia, Benjamin Haibe-Kains, Mathieu Lupien
The fractured landscape of RNA-seq alignment: The default in our STARs. Sara Ballouz, Alexander Dobin, Thomas R Gingeras, Jesse Gillis
miRAW: A deep learning approach to predict miRNA targets by analyzing whole miRNA transcripts. Albert Pla Planas, Xiangfu Zhong, Simon Rayner
Classifying cells with Scasat – a tool to analyse single-cell ATAC-seq. Syed Murtuza Baker, Connor Rogerson, Andrew Hayes, Andrew D. Sharrocks, Magnus Rattray
scmap – A tool for unsupervised projection of single cell RNA-seq data. Vladimir Yu. Kiselev, Andrew Yiu, Martin Hemberg
Expressed Exome Capture Sequencing (EecSeq): a method for cost-effective exome sequencing for all organisms with or without genomic resources. Jonathan B Puritz, Kathleen E Lotterhos
Tn5Prime, a Tn5 based 5′ Capture Method for Single Cell RNA-seq. Charles Cole, Ashley Byrne, Anna E. Beaudin, E. Camilla Forsberg, Christopher Vollmers
Mantis: A Fast, Small, and Exact Large-Scale Sequence Search Index. Prashant Pandey, Fatemeh Almodaresi, Michael A. Bender, Michael Ferdman, Rob Johnson, Rob Patro
SpatialDE – Identification of spatially variable genes. Valentine Svensson, Sarah A. Teichmann, Oliver Stegle
Technology-independent estimation of cell type composition using differentially methylated regions. Stephanie C. Hicks, Rafael A. Irizarry
Revisiting criteria for plant miRNA annotation in the era of big data. Michael J. Axtell, Blake C. Meyers
OMGene: Mutual improvement of gene models through optimisation of evolutionary conservation. Michael Dunne, Steven Kelly
Research practice
Community-driven data analysis training for biology. Bérénice Batut, Saskia Hiltemann, Andrea Bagnacani, Dannon Baker, Vivek Bhardwaj, Clemens Blank, Anthony Bretaudeau, Loraine Guéguen, Martin Čech, John Chilton, Dave Clements, Olivia Doppelt-Azeroual, Anika Erxleben, Mallory Freeberg, Simon Gladman, Youri Hoogstrate, Hans-Rudolf Hotz, Torsten Houwaart, Pratik Jagtap, Delphine Lariviere, Gildas Le Corguillé, Thomas Manke, Fabien Mareuil, Fidel Ramírez, Devon Ryan, Florian Sigloch, Nicola Soranzo, Joachim Wolff, Pavankumar Videm, Markus Wolfien, Aisanjiang Wubuli, Dilmurat Yusuf, Rolf Backofen, Anton Nekrutenko, Björn Grüning
How Changes in Common Practice Can Improve the Quality of Biomedical Science. Matthias Steinfath, Silvia Vogl, Norman Violet, Franziska Schwarz, Hans Mielke, Thomas Selhorst, Matthias Greiner, Gilbert Schoenfelder
The impact factor fallacy. Frieder Michel Paulus, Nicole Cruz, Soeren Krach
Explanation implies causation? Leslie Myint, Jeffrey T. Leek, Leah R. Jager
Assessing value of biomedical digital repositories. Chun-Nan Hsu, Anita Bandrowski, Jeffrey S. Grethe, Maryann E. Martone
Manipulating the alpha level cannot cure significance testing – comments on “Redefine statistical significance”. Trafimow D, Amrhein V, Areshenkoff CN, Barrera-Causil C, Beh EJ, Bilgiç Y, Bono R, Bradley MT, Briggs WM, Cepeda-Freyre HA, Chaigneau SE, Ciocca DR, Carlos Correa J, Cousineau D, de Boer MR, Dhar SS, Dolgov I, Gómez-Benito J, Grendar M, Grice J, Guerrero-Gimenez ME, Gutiérrez A, Huedo-Medina TB, Jaffe K, Janyan A, Karimnezhad A, Korner-Nievergelt F, Kosugi K, Lachmair M, Ledesma R, Limongi R, Liuzza MT, Lombardo R, Marks M, Meinlschmidt G, Nalborczyk L, Nguyen HT, Ospina R, Perezgonzalez JD, Pfister R, Rahona JJ, Rodríguez-Medina DA, Romão X, Ruiz-Fernández S, Suarez I, Tegethoff M, Tejo M, van de Schoot R, Vankov I, Velasco-Forero S, Wang T, Yamada Y, Zoppino FC, Marmolejo-Ramos F.
Key steps to avoiding artistry with significance tests. C Patrick Doncaster, Thomas H G Ezard
If funders and libraries subscribed to open access: The case of eLife, PLOS, and BioOne. Willinsky J, Rusk M.
Oxford Nanopore sequencing in a research-based undergraduate course. Yi Zeng, Christopher Herbert Martin
Why not…
Dietary and behavioral inferences from dental pathology and non-masticatory wear on dentitions from a British medieval town. Ian Towle, Carole Davenport, Joel Irish, Isabelle De Groote
Cobwebbing in the city: urbanization-driven changes in web-building are decoupled from body size in an orb-web spider. Maxime Dahirel, Maarten De Cock, Pieter Vantieghem, Dries Bonte


 (No Ratings Yet)
(No Ratings Yet)
 (2 votes)
(2 votes)


 (3 votes)
(3 votes)