This interview first featured in Development.
Nipam Patel is a developmental biologist based at the University of California, Berkeley, USA, where he uses a variety of organisms to study the evolution of developmental systems, from arthropod body plans to butterfly colouration. We asked him about his career and scientific interests, his role as an editor at Development, and his growing butterfly collection.

When did you first become interested in developmental biology?
As a high school student. The high school I went to was a public school in Texas with an amazing faculty. I actually had a course in developmental biology then, mainly vertebrate embryology taught out of an ancient little German textbook. Our teacher, Rayburn Ray, also had us doing lab experiments: he would get us chicken eggs to do embryology with. Between my junior and senior high school years I was in a summer programme at UT Austin and got into a lab that did blood cell development work in chickens, where I got to design my own project. So, in fact, my first two papers were from the work I did on chick development as a high school student. These experiences really got me hooked on developmental biology. Then, when I went to college, I was fortunate enough to go into Malcolm Steinberg’s lab as soon as I started as a freshman. From that point on, I knew I was either going to be a developmental biologist or an immunologist, and in the end developmental biology won.
During your PhD you worked on Drosophila, but have since moved on to a variety of other organisms. Why?
I did my PhD with Corey Goodman (Stanford University) at the time when the lab had people working on both grasshoppers and Drosophila. I started off working only on Drosophila, but by the time I left I was one of the few people working on grasshoppers in addition to a lot of other odd creatures. A fortuitous experiment led to my finding that a monoclonal antibody (made in collaboration with Thomas Kornberg’s lab) was cross-reactive to Engrailed in a variety of species. This is what got me really excited about evo devo – I realised that, with reagents like this, many of those experiments in comparative developmental biology that people had been talking about could be done relatively quickly. We still do a little Drosophila work, but it is very much the minority.
What are the challenges and benefits of working with non-canonical organisms (and establishing new systems, like you recently did with Parhyale)?
The downside is that you can’t just order antibodies for the protein you want to work on; reagents aren’t easy to come by. On the other hand, everything you do and discover is new, and you are not competing against a dozen other labs doing the exact same experiment. It is a lot of fun, but it is challenging. You need to have a certain mindset to be willing to spend a lot of time developing very basic techniques. But it pays off. After all the work that we and others in the Parhyale community have done, the techniques we can now use in Parhyale are pretty impressive (although it’s obviously still orders of magnitude more work than doing experiments in flies). Also, these days many of the techniques being developed in traditional model systems can be quickly shifted to other species. We specifically pick models that provide both evolutionary insights and have fascinating attributes that make them stand out from standard genetic model systems. There is no perfect system though. For example, Parhyale for us has a lot of wonderful properties, but one downside is that its genome is 10% bigger than the human genome. So it took a while before sequencing costs dropped enough to make it feasible for the community to tackle its genome.
What are the scientific questions that interest you at the moment?
We are working in three main areas. One is the evolution of the body plan, currently focusing on the function of Hox genes. Although these genes have been studied for a long time, there are still many open questions in animals outside of Drosophila – and now we can do functional experiments, for example using CRISPR/Cas9. We have been focused on understanding Hox function primarily in Parhyale, but complementing this with studies in other crustaceans and insects to better understand their role in evolution. A few years ago we also made the accidental discovery that Parhyale has the ability to replace the germline in a way that seems very different to most other animals, so this is another area of work in the lab.
Our newest area of work grows out of my butterfly collecting hobby, which started when I was 8 years old. I have always tried to do some butterfly work myself, and some people might be familiar with my work on gynandromorphs. Now, I finally have students in the lab whose projects are to study butterflies. We are really interested in structural colours. Many of the colours that you see in butterflies, like reds, yellows and browns, are generated from pigments. Other colours, especially greens and blues, are often structural, created by nanostructures of the scales. These colours have evolved over and over again, so there are many different solutions that can be found, and there are hundreds of beautiful papers from optical physicists that have worked out the physics of these structures. But, in most cases, when the physicists figure out how a particular case works they move on to try to make a man-made material that has the same properties, rather than work out how the butterfly builds these structures during development. This is a very new area of work in the lab, but it is something that I am personally very excited about.
Do you have a personal butterfly collection?
I do! I actually have a sizable butterfly collection. I started when I was young, and during my teenage years I collected throughout the USA, India, Africa and Europe. More recently I have been buying some old collections, so I have quite a sizeable collection of my own. This has actually proved really useful for our own experiments. For example, one of the structural colours we work on is in a group of butterflies called the Achillides swallowtails. There are 25 species, and I have 24 of them in my collection. So, if I want to look at the scales of one of the species I don’t even have to go to a museum. When I travel now, I photograph them instead of collecting, and this has also been a great way to learn more about their life histories.
Your research has a strong visual component. How important is aesthetics to your research?
It is very important to me. Our lab has a reputation for generating high-quality images. I have a high standard and I hope that my students and postdocs do too. When people look at our figures I want them to see things clearly, and not have to just take our word for details we claim to show. Often, what we strive for in our images is to have them look as good as the actual preparation, because we take great efforts to make our preparations as high quality as we can. But I think this is one of the beauties of the whole field of developmental biology. It is a visually stunning area to work in, and the images have a lot of impact and carry an incredible amount of information. You can convey what you are working on really simply with a couple of images. I have also always been into nature photography, and the photographic qualities of imaging really drive the aesthetic appeal for me.
What are the challenges of quantitative imaging?
One issue is to know what to quantitate. Nowadays you can take many images and extract a lot of numerical information, but I am not always sure it is useful. Another challenge that has become really apparent is how to capture complex spatial information. Quantitating level or timing is relatively easy, but finding automated and quantitative ways to capture patterns, for example, is a lot harder. Take the work I do on gynandromorphs. I can easily look at the wings and immediately tell you where the boundaries are, as I know the butterflies very well, but teaching a computer how to do it is a huge challenge. More and more data are in the form of images on the internet. How to mine those data easily is a different type of quantitative imaging problem.
In the last few years funding bodies have put an increased emphasis on translational impact. Do you think the evo devo field has been particularly affected by this?
Partially yes. I think a lot of evo devo people didn’t necessarily get NIH funding anyway, so the move towards translational research most impacted those that did. Traditionally, a lot of evo devo funding has come from the NSF, which is much more open to that kind of work. However, evo devo has been affected by the general tightness of funding. This, in addition to there being an increasing number of people in the field, has made it harder to secure funding.
You have been an editor with Development for several years. How has your experience as an editor been?
It has been really fulfilling. First of all it allows me to keep up with the leading edge of the evo devo field, not only through the papers that are submitted but also because, as an editor, people often approach me about whether the work they are doing is appropriate for Development. I have enjoyed trying to help out authors who have interesting stories to tell, and making sure their work gets published. It has also been fun to handle papers from more standard systems, such as Drosophila and sea urchins.
Is there any particular type of paper or topics that you would like to see more of in Development?
Development has published many of the very classic evo devo papers since the early days of the field, including some of mine when I was a graduate student. I would like to see us attract those very best evo devo papers in greater numbers, high-quality papers that have compelling stories to tell. The journal has very high standards, so they have to be very high-quality papers. In general, what might have been published 5 or 10 years ago isn’t going to pass the bar nowadays. Increasingly, there is the ability to conduct manipulative experiments in non-canonical systems, so in many cases we would expect it to some degree. There are, of course, exceptions, and a particularly striking descriptive paper may still be appropriate.
There is often this notion that Development is only a journal for mechanistic development, which some people interpret as only molecular mechanism. I want to make it clear that this is not the case. Those papers that have really important implications for evolutionary mechanisms are also very welcome in Development.
Do you have any advice for young scientists?
I think it is unfortunate how senior people sometimes start off by talking about how funding is hard. There have always been challenges. I think that if young people are very excited and passionate about what they work on, they should go for it. It is tempting to think ‘I am going to work on this because it is a hot field with a future’. This is very hard to predict, and in 6 or 7 years time it might be completely transformed and not the hot field anymore. I think that if you have a passion for something and you think you can find novel, interesting ways to approach it, that is really what you should do.
 (5 votes)
(5 votes)
 Loading...
Loading...


 (2 votes)
(2 votes) (No Ratings Yet)
(No Ratings Yet)

 uthors present a mouse model for tuberous sclerosis complex in which the gene Tsc1 is ablated from eye-progenitor cells, leading to the classic hallmarks of the disease. This demonstrates a role for Tsc1 in regulating several aspects of the development of the visual-pathway. Read the paper
uthors present a mouse model for tuberous sclerosis complex in which the gene Tsc1 is ablated from eye-progenitor cells, leading to the classic hallmarks of the disease. This demonstrates a role for Tsc1 in regulating several aspects of the development of the visual-pathway. Read the paper 

 Kim, Parrish and colleagues show that Rop forms a complex with the exocyst, and that this complex predominates in primary over terminal dendrites. Membrane-associated proteins, on the other hand, preferentially diffuse from primary dendrites into terminal ones, suggesting that diffusion supplies membranous material for terminal dendritic growth. Read the paper
Kim, Parrish and colleagues show that Rop forms a complex with the exocyst, and that this complex predominates in primary over terminal dendrites. Membrane-associated proteins, on the other hand, preferentially diffuse from primary dendrites into terminal ones, suggesting that diffusion supplies membranous material for terminal dendritic growth. Read the paper  ErB2 and ErB3 are two receptor tyrosine kinases expressed in the extravillous trophoblast (EVT) lineage in the placenta. Fock and colleagues demonstrate that neuregulin 1 (NRG1) promotes EVT formation and suppresses trophoblast apoptosis through the activation of ErB2 and ErB3. Read the paper
ErB2 and ErB3 are two receptor tyrosine kinases expressed in the extravillous trophoblast (EVT) lineage in the placenta. Fock and colleagues demonstrate that neuregulin 1 (NRG1) promotes EVT formation and suppresses trophoblast apoptosis through the activation of ErB2 and ErB3. Read the paper  Luman is an ER transmembrane protein that can undergo proteolysis and whose N-terminal fragment can act as a transcription factor. This study shows that Luman can regulate expression, localisation and stability of DC-STAMP, a downstream effector of RANKL, the protein that initiates osteoclastogenesis; making Luman a regulator of osteoclast differentiation. Read the paper
Luman is an ER transmembrane protein that can undergo proteolysis and whose N-terminal fragment can act as a transcription factor. This study shows that Luman can regulate expression, localisation and stability of DC-STAMP, a downstream effector of RANKL, the protein that initiates osteoclastogenesis; making Luman a regulator of osteoclast differentiation. Read the paper  Hall and Verheyen show that Dsor1, a Drosophila homolog of MEK that is activated by Ras, promotes transcription of Wg target genes by interacting with Armadillo and preventing its degradation. Ras-Dsor1 activity seems to be mediated by the insulin-like growth factor receptor. Read the paper
Hall and Verheyen show that Dsor1, a Drosophila homolog of MEK that is activated by Ras, promotes transcription of Wg target genes by interacting with Armadillo and preventing its degradation. Ras-Dsor1 activity seems to be mediated by the insulin-like growth factor receptor. Read the paper 
 a and colleagues investigated the spawning behaviour of the Japanese flying squid, comparing eggs spawned in a tank with a temperature gradient with eggs spawned in a tank without a temperature gradient. Only eggs spawned in the tank with the temperature gradient survived. Paralarvae survived for ten days, allowing observation of advanced stage paralarvae. Read the paper
a and colleagues investigated the spawning behaviour of the Japanese flying squid, comparing eggs spawned in a tank with a temperature gradient with eggs spawned in a tank without a temperature gradient. Only eggs spawned in the tank with the temperature gradient survived. Paralarvae survived for ten days, allowing observation of advanced stage paralarvae. Read the paper 





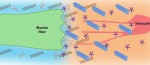 Tendons are ECM-rich structures that interconnect muscles and bones. Here, Subramanian and Schilling review how intrinsic mechanisms as well as extrinsic factors, such as mechanical force, regulate the ECM to control tendon development and maturation. See the Review on p.
Tendons are ECM-rich structures that interconnect muscles and bones. Here, Subramanian and Schilling review how intrinsic mechanisms as well as extrinsic factors, such as mechanical force, regulate the ECM to control tendon development and maturation. See the Review on p.