In Development this week (Vol. 142, Issue 10)
Posted by Seema Grewal, on 12 May 2015
Here are the highlights from the current issue of Development:
Capping off sesamoid bone development
 Sesamoid bones are small, flat bones that are embedded within tendons. To date, it has been thought that these bones develop within tendons in response to mechanical signals, but now (on p. 1831) Elazar Zelzer and colleagues challenge this assumption, focussing on the patella (the kneecap), which is the largest sesamoid bone in the human body. They show that that, in mice, the patella initially develops as a bony process that is part of the adjacent femur bone. Subsequently, they report, the patella is separated from the femur during the process of joint formation. This process is regulated by mechanical load; patella separation is perturbed in mutant embryos that are devoid of contracting muscle. The authors further demonstrate that, similar to bone eminences – superstructures that mediate bone-tendon attachment – the patella arises from progenitors that express the chondrocyte marker Sox9 and the tendon marker scleraxis (Scx) and that are regulated by TGFβ/BMP signalling. Together, these findings provide a new model for patella development and highlight that a high degree of plasticity exists during skeletal patterning and development.
Sesamoid bones are small, flat bones that are embedded within tendons. To date, it has been thought that these bones develop within tendons in response to mechanical signals, but now (on p. 1831) Elazar Zelzer and colleagues challenge this assumption, focussing on the patella (the kneecap), which is the largest sesamoid bone in the human body. They show that that, in mice, the patella initially develops as a bony process that is part of the adjacent femur bone. Subsequently, they report, the patella is separated from the femur during the process of joint formation. This process is regulated by mechanical load; patella separation is perturbed in mutant embryos that are devoid of contracting muscle. The authors further demonstrate that, similar to bone eminences – superstructures that mediate bone-tendon attachment – the patella arises from progenitors that express the chondrocyte marker Sox9 and the tendon marker scleraxis (Scx) and that are regulated by TGFβ/BMP signalling. Together, these findings provide a new model for patella development and highlight that a high degree of plasticity exists during skeletal patterning and development.
FGF receptors sculpt synaptogenesis
 A balanced network of excitatory and inhibitory synapses is required for correct brain function, and any perturbations to this balance can give rise to neurological and psychiatric disorders. It has been shown previously that FGF22 and FGF7 promote excitatory or inhibitory synapse formation, respectively, in the hippocampus, but how do these ligands mediate their synaptogenic effects? Here, Hisashi Umemori and co-workers use various FGF receptor knockout mice to address this question (p. 1818). They first show that excitatory presynaptic differentiation is impaired inFgfr2b and Fgfr1b mutant mice. Following on from this, they reveal that both FGFR2b and FGFR1b act downstream of FGF22 and are required for FGF22-dependent excitatory presynaptic differentiation. The authors further show that the kinase activity of FGFR2b as well as its ability to bind to FRS2 and PI3K is required for it to respond to FGF22. By contrast, they report, inhibitory presynaptic differentiation is defective only in Fgfr2b, and not Fgfr1b, mutants. In line with this, they demonstrate that FGF7 requires FGFR2b and not FGFR1b to mediate its effect on inhibitory presynaptic differentiation. Together, these findings indicate that distinct but overlapping sets of FGF receptors sculpt excitatory and inhibitory synapse formation in the mammalian brain.
A balanced network of excitatory and inhibitory synapses is required for correct brain function, and any perturbations to this balance can give rise to neurological and psychiatric disorders. It has been shown previously that FGF22 and FGF7 promote excitatory or inhibitory synapse formation, respectively, in the hippocampus, but how do these ligands mediate their synaptogenic effects? Here, Hisashi Umemori and co-workers use various FGF receptor knockout mice to address this question (p. 1818). They first show that excitatory presynaptic differentiation is impaired inFgfr2b and Fgfr1b mutant mice. Following on from this, they reveal that both FGFR2b and FGFR1b act downstream of FGF22 and are required for FGF22-dependent excitatory presynaptic differentiation. The authors further show that the kinase activity of FGFR2b as well as its ability to bind to FRS2 and PI3K is required for it to respond to FGF22. By contrast, they report, inhibitory presynaptic differentiation is defective only in Fgfr2b, and not Fgfr1b, mutants. In line with this, they demonstrate that FGF7 requires FGFR2b and not FGFR1b to mediate its effect on inhibitory presynaptic differentiation. Together, these findings indicate that distinct but overlapping sets of FGF receptors sculpt excitatory and inhibitory synapse formation in the mammalian brain.
ATRX: keeping quiet in the embryo
 Human embryos are particularly susceptible to chromosome instability (CIN) and errors in chromosome segregation, but the molecular mechanisms that regulate and sense CIN in mammalian embryos are unclear. Here, on p. 1806, Maria Viveiros and colleagues investigate the role of the chromatin remodelling protein ATRX in early mouse embryos. They first show that ATRX, which is transmitted to the early zygote through the maternal germ line, localises to pericentric heterochromatin (PCH) within the maternal pronucleus, where it is required for the transcriptional repression of major satellite transcripts. The loss of ATRX hence leads to the abnormal expression of maternal satellite transcripts. The authors also demonstrate that the maternal inheritance of ATRX helps to set up an epigenetic asymmetry between the maternal and paternal chromosomes, which might be implicated in facilitating chromosome segregation. In line with this, ATRX loss, they report, causes abnormal centromeric mitotic recombination and an increase in double-strand DNA breaks. Overall, these data highlight an important role for ATRX in the early mouse embryo and provide new insights into how CIN is controlled in early mammalian development.
Human embryos are particularly susceptible to chromosome instability (CIN) and errors in chromosome segregation, but the molecular mechanisms that regulate and sense CIN in mammalian embryos are unclear. Here, on p. 1806, Maria Viveiros and colleagues investigate the role of the chromatin remodelling protein ATRX in early mouse embryos. They first show that ATRX, which is transmitted to the early zygote through the maternal germ line, localises to pericentric heterochromatin (PCH) within the maternal pronucleus, where it is required for the transcriptional repression of major satellite transcripts. The loss of ATRX hence leads to the abnormal expression of maternal satellite transcripts. The authors also demonstrate that the maternal inheritance of ATRX helps to set up an epigenetic asymmetry between the maternal and paternal chromosomes, which might be implicated in facilitating chromosome segregation. In line with this, ATRX loss, they report, causes abnormal centromeric mitotic recombination and an increase in double-strand DNA breaks. Overall, these data highlight an important role for ATRX in the early mouse embryo and provide new insights into how CIN is controlled in early mammalian development.
Divide but stay together: cytokinesis in tubes
 During development, epithelial tubes often need to grow while still maintaining their barrier properties. How can cells divide without disrupting the integrity of the tubular epithelium? On p. 1794, Markus Affolter and colleagues address this question in the Drosophila larval tracheal system. In a particular subset of tracheal tubes, there is extensive remodelling during the early third instar, such that unicellular tubes, in which a single cell encircles the lumen and creates junctions with itself, transform into multicellular tubes, a process accompanied by proliferation. The authors demonstrate that this transition involves cell intercalation to replace the autocellular junctions with intercellular ones. Depending on cell length, mitosis may occur either before or after this junctional remodelling is complete, thus generating two major classes of cytokinesis events. In both cases, cytokinesis is asymmetric, with the new membrane extending from the side of the cell where the nucleus is located. In rare cases, this can lead to the formation of a binucleate and an anucleate daughter. The authors further find that Dpp signalling is required for appropriate junctional remodelling and cell division. Together, these data provide insights into how barrier integrity can be maintained through cell division in these tubular structures.
During development, epithelial tubes often need to grow while still maintaining their barrier properties. How can cells divide without disrupting the integrity of the tubular epithelium? On p. 1794, Markus Affolter and colleagues address this question in the Drosophila larval tracheal system. In a particular subset of tracheal tubes, there is extensive remodelling during the early third instar, such that unicellular tubes, in which a single cell encircles the lumen and creates junctions with itself, transform into multicellular tubes, a process accompanied by proliferation. The authors demonstrate that this transition involves cell intercalation to replace the autocellular junctions with intercellular ones. Depending on cell length, mitosis may occur either before or after this junctional remodelling is complete, thus generating two major classes of cytokinesis events. In both cases, cytokinesis is asymmetric, with the new membrane extending from the side of the cell where the nucleus is located. In rare cases, this can lead to the formation of a binucleate and an anucleate daughter. The authors further find that Dpp signalling is required for appropriate junctional remodelling and cell division. Together, these data provide insights into how barrier integrity can be maintained through cell division in these tubular structures.
A double take on the segmentation clock
 The segmentation clock, which controls the periodic formation of somites along the vertebrate body axis, involves the oscillating expression of clock genes in presomitic mesoderm (PSM) cells. Oscillations are synchronised between cells, giving rise to a sweeping wave of gene expression throughout the PSM. This cyclic wave of gene expression is known to slow as it moves anteriorly, but the causes and implications of this slowing have remained unclear. Here, Sharon Amacher and co-workers investigate segmentation clock dynamics in zebrafish embryos (p. 1785). Using a her1:her1-venus reporter to visualise clock gene oscillations in real-time, the authors show that the periodicity of oscillations slows as PSM cells become displaced anteriorly. This slowing gives rise to a situation in which cells that are one somite apart are actually in opposite phases of the clock. Thus, they report, a one-segment periodicity is observed in the posterior of the embryo, whereas a two-segment spatial periodicity is seen in the anterior. The researchers further demonstrate that neighbouring cells oscillate synchronously in both the posterior and anterior PSM until they are incorporated into somites. Based on these findings, the authors propose an updated model of the segmentation clock.
The segmentation clock, which controls the periodic formation of somites along the vertebrate body axis, involves the oscillating expression of clock genes in presomitic mesoderm (PSM) cells. Oscillations are synchronised between cells, giving rise to a sweeping wave of gene expression throughout the PSM. This cyclic wave of gene expression is known to slow as it moves anteriorly, but the causes and implications of this slowing have remained unclear. Here, Sharon Amacher and co-workers investigate segmentation clock dynamics in zebrafish embryos (p. 1785). Using a her1:her1-venus reporter to visualise clock gene oscillations in real-time, the authors show that the periodicity of oscillations slows as PSM cells become displaced anteriorly. This slowing gives rise to a situation in which cells that are one somite apart are actually in opposite phases of the clock. Thus, they report, a one-segment periodicity is observed in the posterior of the embryo, whereas a two-segment spatial periodicity is seen in the anterior. The researchers further demonstrate that neighbouring cells oscillate synchronously in both the posterior and anterior PSM until they are incorporated into somites. Based on these findings, the authors propose an updated model of the segmentation clock.
PLUS:
An interview with Juergen Knoblich
 Juergen Knoblich is a senior scientist and deputy scientific director of the Institute of Molecular Biotechnology of the Austrian Academy of Sciences in Vienna. We met Juergen at the 56th Annual Drosophila Research Conference, where we asked him about his work in this model system and, more recently, on human cerebral organoids, and about his thoughts on recent technological developments and the funding situation.
Juergen Knoblich is a senior scientist and deputy scientific director of the Institute of Molecular Biotechnology of the Austrian Academy of Sciences in Vienna. We met Juergen at the 56th Annual Drosophila Research Conference, where we asked him about his work in this model system and, more recently, on human cerebral organoids, and about his thoughts on recent technological developments and the funding situation.
Hematopoietic development at high altitude: blood stem cells put to the test
 In February 2015, scientists gathered for the Keystone Hematopoiesis meeting, which was held at the scenic Keystone Resort in Colorado, USA. During the exciting program, field leaders and new investigators presented discoveries that spanned developmental and adult hematopoiesis within both physiologic and pathologic contexts. See the Meeting Review on p. 1728
In February 2015, scientists gathered for the Keystone Hematopoiesis meeting, which was held at the scenic Keystone Resort in Colorado, USA. During the exciting program, field leaders and new investigators presented discoveries that spanned developmental and adult hematopoiesis within both physiologic and pathologic contexts. See the Meeting Review on p. 1728
Building the backbone: the development and evolution of vertebral patterning
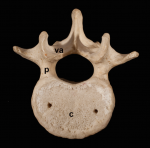 The segmented vertebral column comprises a repeat series of vertebrae, each consisting of the vertebral body and the vertebral arches. Despite being a defining feature of vertebrates, much remains to be understood about vertebral development and evolution. Particular controversy surrounds whether vertebral structures are homologous across vertebrates, how somite and vertebral patterning are connected, and the developmental origin of vertebral bone-mineralizing cells. Here, Roger Keynes and colleagues consider these issues. See the Review on p. 1733
The segmented vertebral column comprises a repeat series of vertebrae, each consisting of the vertebral body and the vertebral arches. Despite being a defining feature of vertebrates, much remains to be understood about vertebral development and evolution. Particular controversy surrounds whether vertebral structures are homologous across vertebrates, how somite and vertebral patterning are connected, and the developmental origin of vertebral bone-mineralizing cells. Here, Roger Keynes and colleagues consider these issues. See the Review on p. 1733


 (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)