I presume the first question that comes into people’s mind when they read the title would be “What are cnidarians?” I also presume a logical follow-up question would be “Why are you working on cnidarians?” If you haven’t asked these questions, I suspect that you are either my colleague and are sitting in the office next to me, or that you too work in the field of evo-devo.
We have all encountered cnidarians primarily in a non-scientific way, probably while swimming somewhere in the sea. And if the exposure was a direct skin-to-skin contact, there is a possibility that it wasn’t pleasant for the human participant. The unpleasant encounters are underpinned by the determining characteristic of cnidarians – the stinging cells (cnidocytes). Cnidarians are a phylum of metazoans, which separated from the lineage that gave rise to bilaterians more than 600 million years ago (Figure 1.).

Figure 1 – Phylogenetic relationship of the extant cnidarian classes
Today’s cnidarians include 5 classes, among which are the well-known sea anemones, corals and jelly fish. They are relatively simple animals, with only two germ layers (endoderm and ectoderm) and a small number of cell types. However, this morphological simplicity is deceptive, as it was recently reported that cnidarians possess a complex genome and a gene regulatory landscape that are comparable to any of the “higher” (although I prefer not to refer to them that way) animals, namely bilaterians. This intricate relationship of simple morphology and a complex genetic toolkit makes cnidarians ideal for studying the emergence of complex traits. The problems which interest the researchers in the lab of Uli Technau at the Department of Molecular Evolution and Development at the University of Vienna, Austria belong to some of the most fundamental questions in biology in general: the evolution of germ layers, left and right symmetry and nervous systems. The researchers in the lab come from different backgrounds – developmental biologists, molecular biologists, geneticists, zoologists. The variety of education of the researchers is what contributes greatly to the diversity of interests and topics.

Figure 2 – The life cycle of Nematostella vectensis
In order to tackle these questions most researchers in the lab use the starlet sea anemone – Nematostella vectensis (Figure 2.). This slowly evolving cnidarian proved itself to be one of the favourite cnidarian models, due to its willingness to be kept and to reproduce under laboratory conditions. Animals can be spawned at request, by using a combination of temperature and light setting changes and therefore provide us with fresh embryos every day. As our interest especially lies in the developmental origins of the previously mentioned biological basics it is crucial that we can access every stage of development easily. The enthusiasm with which Nematostella spawns in the lab has encouraged the development of techniques to work with it. The development of gene interference systems and transgenesis made work with these organisms much easier, but they still present somewhat of a challenge, because in order to close the life cycle (i.e. get the F1 generation), one must wait for 4-6 months (Figure 2). So do not expect quick results while working in the cnidarian field, but expect them to be very pretty…
A typical day in the cnidarian life for the researchers starts with coffee (I guess like in most research institutions all over the world), and for the animals with feeding and cleaning, and, for the chosen ones on that day, spawning. Spawning is induced, as mentioned before, with the combination of temperature and light change. The chosen animals are carefully washed the day before the spawning and put in incubators, which are timed to change the temperature and light. Usually the animals are kept in the dark at 18˚C, but the temperature in the incubators is 25˚C, and the light is on. We keep our animals in separate boxes for males and females, as to avoid any unplanned fertilization (very rarely the animals will spawn un-induced). After taking the animals out of the incubator, in about an hour to two they produce gametes. The females expel egg packages (eggs surrounded by a thick layer of jelly), and males release sperm into the water. Egg packages are transferred from female boxes into males ones and fertilization happens while you’re finishing your coffee. The embryos are collected, dejellied and one can then use them for any purpose. A very common technique that we subject our embryos to is microinjection. This is used to deliver all sorts of genetic goodies to the embryo: morpholinos, over-expression constructs, transgenic constructs. Most of us in the lab have spent several months trying to master this technique as it can be quite tricky. Especially since it is done in a relatively cold room (13-15˚C), which is not the most comfortable temperature for an almost motionless human being. But once you get there, it is quite satisfying (and maybe prepares you for a future job in human in vitro fertilization).

Figure 3 – A semi-automatic culture system for Nematostella vectensis
After fertilization (and potential microinjection), the eggs cleave and in around a day the embryos start gastrulating. After around a day and a half, the gastrula develops into a swimming larva called planula. After several days of swimming, the planulae settle down on the substrate to metamorphose into primary polyps, which develop 4 tentacles. The polyps start feeding, grow and develop more tentacles. The animals grows continuously and reaches sexual maturity around 4-6 months. Another interesting fact about Nematostella is that it is probably a very long lived animal with little signs of aging. As no detailed research has been done on this problem, there are a lot of open question left to address. Our animals are kept in a semi-automatic culture system in which the quality of the water is under central control (Figure 3., 4. and 5.).

Figure 4 – Nematostella in its home in the culture box.

Figure 5. – Female Nematostella just before laying eggs.
All of the animals are taken care of by our animal technicians and are fed Artemia shrimps (like zebrafish) – hence the slightly pink colour of the Nematostella polyps. They also take care that the animals are clean and that they do not overcrowd their boxes (since they are fond of their asexual reproduction, that does happen from time to time). In addition to Nematostella, our lab houses several other cnidarian species. The ones I believe would draw the most attention are the medusa stages of Clytia hemispherica (a hydrozoan) and Aurelia aurita (a scyphozoan). Aurelia aurita (Video) or the moon jelly is found in most of the world’s seas and you might have seen it live. But don’t worry, these guys don’t sting. The reason why they are so interesting, except for being exceptionally beautiful, is that they have a very elaborate life cycle. Medusozoans in general have a life cycle which alternates between a polyp and a medusa stage (although this typical life cycle underwent many changes during evolution, and some lineages completely lost either the medusa, or the polyp stage). The relationship and developmental dynamics of these life stages is also a question that interests some of the researchers in our lab. The lab houses some other cnidarian stages from time to time, usually only to obtain DNA or RNA from the poor things, as establishing a protocol for maintaining a new species in the lab, and especially for closing its life cycle is a hard and lengthy process. When I decided to work on cnidarians, I was attracted to the possibilities that these animals provide to study the characteristics of the ancestor of our lineages. They have been full of surprises and have shown us that this ancient group was much more complex than previously thought. So, whenever a cnidarian biologist has a tough time (which does happen quite often, unfortunately, as they tend to be uncooperative as any other biological system), it is enough to go and see your animals peacefully sitting or floating in water and their almost meditative influence will for sure ease a difficult day. Or just go and have a beer, there is always one in the fridge…
Young Aurelia aureate medusae. At the bottom of the dish ephyrae (“baby medusae”) can be seen.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
 (9 votes)
(9 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)

 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series  (15 votes)
(15 votes)










 The nervous system of bilaterians arises from a small pool of neural progenitor cells (NPCs) that expressesSoxB genes, a family of transcription factors crucial for neurogenesis. The existence of NPCs has thus far been described in diverse species of Bilateria but not in its sister group, the Cnidaria. Hence, the evolutionary origin of NPCs remains obscure. Gemma Richards and Fabian Rentzsch (p.
The nervous system of bilaterians arises from a small pool of neural progenitor cells (NPCs) that expressesSoxB genes, a family of transcription factors crucial for neurogenesis. The existence of NPCs has thus far been described in diverse species of Bilateria but not in its sister group, the Cnidaria. Hence, the evolutionary origin of NPCs remains obscure. Gemma Richards and Fabian Rentzsch (p.  Continuous neurogenesis in the adult hippocampus is achieved by a tightly regulated balance between adult neural stem cell (aNSCs) self-renewal and differentiation. aNSC self-renewal is maintained by the action of ‘stemness’ genes, including Notch. Conversely, aNSC differentiation involves both the inactivation of the ‘stemness’ genes and activation of pro-neural genes. Adult hippocampal neurogenesis is regulated by intrinsic stimuli such as epigenetic modifications, as well as extrinsic inputs such as exercise, diet or hypoxia, which ultimately cause metabolic stress. However, the molecular mechanisms linking metabolic changes to the epigenetic control of aNSCs remain unclear. Using genetic ablation and pharmacological manipulation in mouse (p.
Continuous neurogenesis in the adult hippocampus is achieved by a tightly regulated balance between adult neural stem cell (aNSCs) self-renewal and differentiation. aNSC self-renewal is maintained by the action of ‘stemness’ genes, including Notch. Conversely, aNSC differentiation involves both the inactivation of the ‘stemness’ genes and activation of pro-neural genes. Adult hippocampal neurogenesis is regulated by intrinsic stimuli such as epigenetic modifications, as well as extrinsic inputs such as exercise, diet or hypoxia, which ultimately cause metabolic stress. However, the molecular mechanisms linking metabolic changes to the epigenetic control of aNSCs remain unclear. Using genetic ablation and pharmacological manipulation in mouse (p.  In both plants and animals, cellular senescence is not only an age-related process, but can also contribute to developmental programs. In plants, senescence can occur with age and in response to suboptimal growing conditions to reallocate nutrients from the leaves to the developing parts of the plant, particularly to maturing seeds. However, the interplay between age- or environmentally induced senescence and developmental programs is still unclear. Using a ChIP-Seq approach in Arabidopsis (p.
In both plants and animals, cellular senescence is not only an age-related process, but can also contribute to developmental programs. In plants, senescence can occur with age and in response to suboptimal growing conditions to reallocate nutrients from the leaves to the developing parts of the plant, particularly to maturing seeds. However, the interplay between age- or environmentally induced senescence and developmental programs is still unclear. Using a ChIP-Seq approach in Arabidopsis (p.  During development, somites form by periodic budding from the pre-somitic mesoderm (PSM), and give rise to the vertebral column and most of the muscles and skin. This process is driven by the pulsatile expression of ‘clock genes’, the expression of which is synchronized across the PSM. This synchronicity is regulated by the Notch pathway. Notch1 and its ligand Delta1 (Dll1) are reported to be expressed in a continuous gradient in the PSM and it is unclear how these static receptor and ligand profiles can drive and synchronise pulsatile gene expression. Through experiments in mouse and chick (p.
During development, somites form by periodic budding from the pre-somitic mesoderm (PSM), and give rise to the vertebral column and most of the muscles and skin. This process is driven by the pulsatile expression of ‘clock genes’, the expression of which is synchronized across the PSM. This synchronicity is regulated by the Notch pathway. Notch1 and its ligand Delta1 (Dll1) are reported to be expressed in a continuous gradient in the PSM and it is unclear how these static receptor and ligand profiles can drive and synchronise pulsatile gene expression. Through experiments in mouse and chick (p.  Building the elaborate neural networks required for brain function involves profound cytoskeleton remodelling during axonal growth and pathfinding. In particular, axonal growth is supported by the growth cone, a dynamic F-actin based structure, and regulated by ADF/Cofilin, an F-actin destabilising protein. Cofilin is activated by dephosphorylation by Slingshot (Ssh), and inhibited by LIMK-mediated phosphorylation. Activity of these regulators is in turn influenced by the small GTPase Rac, which acts via Pak to promote LIMK activity and inhibit Cofilin, but also via a Pak-independent, non-canonical pathway to promote Cofilin activity. However, the molecular mediators of the non-canonical pathway are currently unknown. Here (p.
Building the elaborate neural networks required for brain function involves profound cytoskeleton remodelling during axonal growth and pathfinding. In particular, axonal growth is supported by the growth cone, a dynamic F-actin based structure, and regulated by ADF/Cofilin, an F-actin destabilising protein. Cofilin is activated by dephosphorylation by Slingshot (Ssh), and inhibited by LIMK-mediated phosphorylation. Activity of these regulators is in turn influenced by the small GTPase Rac, which acts via Pak to promote LIMK activity and inhibit Cofilin, but also via a Pak-independent, non-canonical pathway to promote Cofilin activity. However, the molecular mediators of the non-canonical pathway are currently unknown. Here (p. 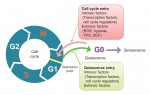 Quiescence has been proposed as a fundamental property of hematopoietic stem cells (HSCs), acting to protect them from functional exhaustion and cellular insults to enable lifelong hematopoietic cell production. Toshio Suda and colleagues review the current methods available to measure quiescence in HSCs and discuss the roles and regulation of HSC quiescence. See the Review on p.
Quiescence has been proposed as a fundamental property of hematopoietic stem cells (HSCs), acting to protect them from functional exhaustion and cellular insults to enable lifelong hematopoietic cell production. Toshio Suda and colleagues review the current methods available to measure quiescence in HSCs and discuss the roles and regulation of HSC quiescence. See the Review on p.  Transcription factors establish the tremendous diversity of cell types in the nervous system by regulating the expression of genes that give a cell its morphological and functional properties. Celine Santiago and Greg Bashaw highlight recent work that has elucidated the functional relationships between transcription factors and the downstream effectors through which they regulate neural connectivity in multiple model systems. See the Review on p.
Transcription factors establish the tremendous diversity of cell types in the nervous system by regulating the expression of genes that give a cell its morphological and functional properties. Celine Santiago and Greg Bashaw highlight recent work that has elucidated the functional relationships between transcription factors and the downstream effectors through which they regulate neural connectivity in multiple model systems. See the Review on p.