Hi!
I am Nuria, a PhD student. I work in the Department of Cell Biology at the University of Santiago de Compostela (Spain).
Our group, the BRAINSHARK group, has been working in the evolutionary changes occurring during the development of the nervous system of a small shark (Scyliorhinus canicula) for many years. Our work mostly focuses on studying the development and the gene expression in this cartilaginous fish comparing with other vertebrates to gain insights in common processes mainly during the early development of the brain, but also in other parts of the nervous system such as the olfactory system or the retina.
When I was an undergrad in Biology, I have never thought that I would work with sharks in a lab! Yes, I know it is a small one, but still a shark! Now, I am using this friendly fish to study the development of the retina.
Sharks as a model for developmental studies
The lesser-spotted dogfish, S. canicula, is an increasingly used model organism for studying embryonic nervous system development.
But why a shark? Chondrichthyans (cartilaginous fishes) constitute a monophyletic group, an ancient lineage of gnathostomous vertebrates characterized by having a cartilaginous endoskeleton, and placoid scales (dermal denticles) covering their body. This group includes two major radiations that diverged over 250 million years ago. Elasmobranchs are one of these groups, including sharks, skates and rays, characterized by having articulated jaws. Its phylogenetic position makes chondrichthyans a key animal group to study the ancestral vertebrate condition of brain organization, because they provide a comparative reference for distinguishing between ancestral and derivative characters (Figure 1). Despite the fact that chondrichthyans represent an ancient vertebrate radiation, they don’t have primitive or unspecialized brain structures. Instead, they present well-developed, large brains, comparable in size to those of birds and mammals. In addition, the lesser-spotted dogfish offers great advantages as (1) the access to a unique collection of genomic resources, (2) the access to the embryonic development in ovo, (3) the possibility of maintaining embryos outside the eggshell for several days, and (4) the extremely slow growth and the big size of embryonic brain, which is really important to make detailed studies of particular regions.
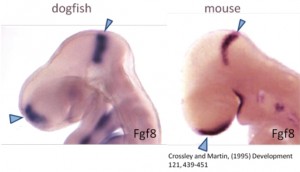 Figure 1. Comparison of a mouse and shark embryo.
Figure 1. Comparison of a mouse and shark embryo.
In sharks, vision, together with other sensory systems such as the lateral line, play important roles in habitat selection and in predatory and reproductive behaviour. Sharks have conspicuous eyes, generally positioned laterally on the head, thus providing some binocular overlap in the visual field (Figure 2).

Figure 2. In sharks the eyes are generally positioned laterally on the head.
As an accessible extension of the brain, the retina offers an exceptional model to extend our knowledge on the development of the nervous system. Wherefore, several morphological and physiological studies have characterized the retinal circuitry and cells types in different elasmobranchs species. Fish retina has been found to be useful for studying retinogenesis as it contains retinal stem cells, which give rise to all cell types throughout the entire life of the animal. Our group has contributed significantly with developmental studies dealing with retinal morphogenesis.
Animal maintenance
Embryos of sharks are supplied by the Marine Biological Station of Roscoff (France). They are very young travelers that arrive to our lab packed in plastic bags. Apart form the eggs, sometimes we receive juveniles kindly provided by the Acuaria of Gijón, O Grove and A Coruña (Spain). Then the embryos from different broods and juveniles are raised in a tank of fresh sea water under standard conditions of temperature (16-18º) and 12:12 hours day/night cycle. They are introduced inside plastic bags to facilitate the acclimatization to their new home. The eggs are easily maintained under laboratory conditions until hatching, and the transparency of the egg shells makes it possible to select the required stage of development. The eggs have tendrils that allow their to attach to a substrate such as corals or seaweed. We use the tendrils to anchor the eggs to floating rods speeding up their development. In oviparous shark species as S. canicula, the embryos get their nourishment from a yolk sac. They may take several months to hatching, facilitating the study of developmental processes (Figure 3).
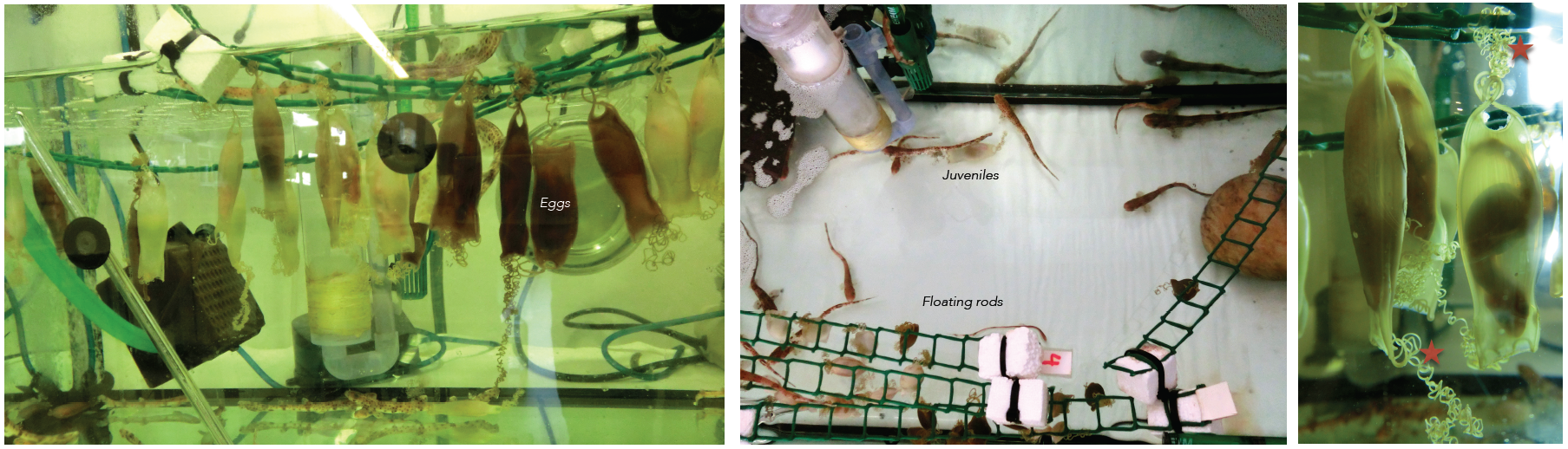
Figure 3. Our shark tanks. Tendrils of the eggs (asterisk). Click to see bigger version.
A typical day in our lab
When we arrive in the morning, we usually do the maintenance of the tank. It is necessary to test the pH, the concentration of nitrites and nitrates, check the temperature, and remove some dirty water and introduce some cleaning new. If we have juveniles, we feed them with frozen shrimp or squid. Finally, we check the embryos one by one, removing embryos appear damaged (when the yolk is broken or when the embryo does not move inside the egg). We can do it because of the transparency of the eggs! This allows us in addition to check the eggs, observe their development without removing the embryo from the egg (the video shows a healthy embryo inside the egg).
Staging embryos
To perform developmental experiments the first thing I have to do is to select the embryos on the stage I need to study. I stage the embryos on the basis of their external features according to Ballard et al. (1993) using a stereoscopic microscope (Figure 4).
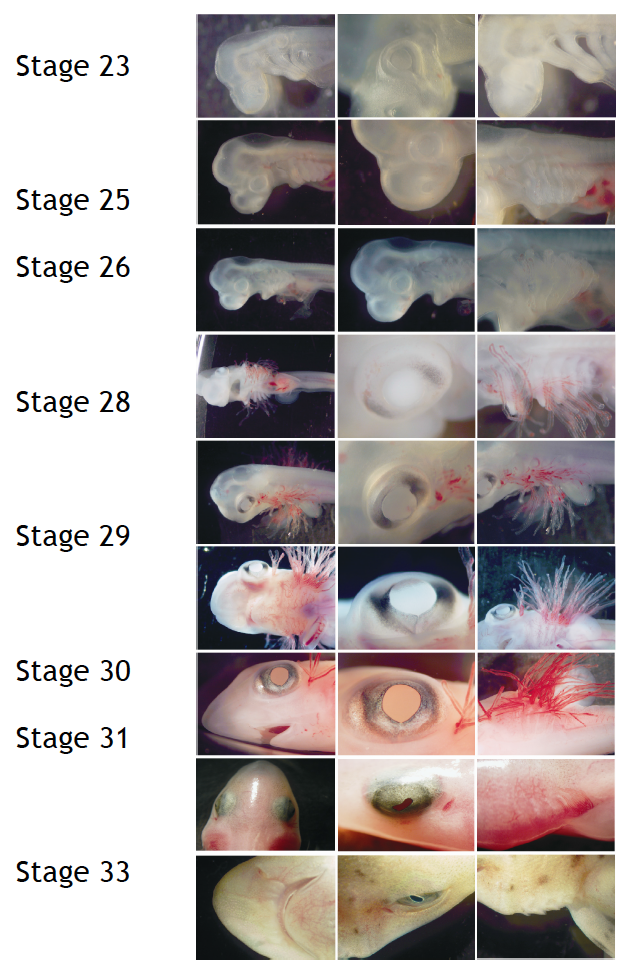
Figure 4. External features during S.canicula development
What do I do with sharks?
Tissue preparation
Little embryos up to stage 32 are deeply anesthetized in seawater commensurate with the adequate measures to minimize animal pain or discomfort. Soon after, I separate the embryo from the yolk before fixation. For embryos from stage 33 onwards and juveniles, I anesthetize in the same way and then, I perfuse their intracardially. Afterward, I remove the eyes and the brain. My colleagues work in other parts of the nervous system, so no rest is wasted (Figure 5).

Figure 5. Tissue preparation. Click to see bigger version.
I usually cut the eyes on a cryostat (sometimes a use a microtome, depending on the further use).
Our group works with the typical developmental techniques such immunohistochemistry, in situ hybridization, tracing, etc. Finally, we study the sections and take the photomicrographs using different types of scopes (Figure 6).
Figure 6. My work place in the lab. and the confocal microscopy station.
Future expectations
We are moving towards a more functional approach, trying to set-up new techniques that allow us to finding molecular determinants that can stabilize the neural stem stage, serve as fate determinants towards the neuronal lineage or can reverse a glial precursor into a neuronal precursor. However genetic manipulation in S. canicula is far from being optimized, it would be the next step to establish testable hypothesis for our descriptive data. To learn the techniques we needed in our lab, I am currently doing a research stay at the Center for Regenerative Therapies (CRTD) of Dresden, founded by Disease Models & Mechanisms (The Company of Biologists).
Acknowledgements: Thanks to Dr. Idoia Quintana to encourage me to write this post and for her kindly review and suggestions, and my college Santiago Pereira for his help in changing the format of the images.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
 (16 votes)
(16 votes)
 Loading...
Loading...



 (1 votes)
(1 votes)

 Malin Parmar heads a research group focused on developmental and regenerative neurobiology at Lund University in Sweden. The ultimate goal of her research is to develop cell therapy for Parkinson’s disease.
Malin Parmar heads a research group focused on developmental and regenerative neurobiology at Lund University in Sweden. The ultimate goal of her research is to develop cell therapy for Parkinson’s disease.

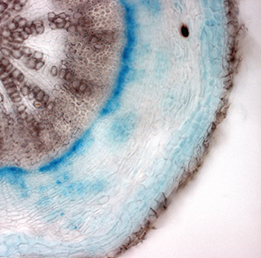 Class I KNOX transcription factors, such as SHOOT MERISTEMLESS (STM) and KNAT1, are known to play a role in the plant shoot apical meristem (SAM), where they are thought to prevent differentiation and hence promote stem cell maintenance. Now, on p.
Class I KNOX transcription factors, such as SHOOT MERISTEMLESS (STM) and KNAT1, are known to play a role in the plant shoot apical meristem (SAM), where they are thought to prevent differentiation and hence promote stem cell maintenance. Now, on p. 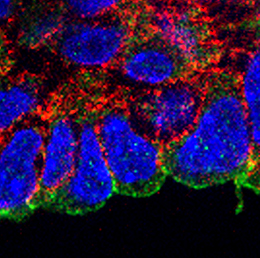 During its development, the heart tube undergoes rapid elongation, fuelled by the addition of cardiac progenitors from the second heart field (SHF). The gene regulatory networks governing SHF formation have been studied extensively, but little is known about the basic cellular features of SHF cells. Now, Robert Kelly and co-workers show that the transcription factor TBX1, which is implicated in both normal SHF development and congenital heart defects, regulates the epithelial properties of mouse SHF cells (p.
During its development, the heart tube undergoes rapid elongation, fuelled by the addition of cardiac progenitors from the second heart field (SHF). The gene regulatory networks governing SHF formation have been studied extensively, but little is known about the basic cellular features of SHF cells. Now, Robert Kelly and co-workers show that the transcription factor TBX1, which is implicated in both normal SHF development and congenital heart defects, regulates the epithelial properties of mouse SHF cells (p. 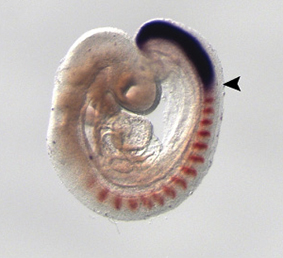 During development, neuromesodermal (NM) stem cells give rise to both neural cells and paraxial presomitic mesoderm (PSM) cells, but what dictates PSM fate? Here, Terry Yamaguchi and colleagues show that a single transcription factor – mesogenin 1 (Msgn1) – acts as a master regulator of PSM development (p.
During development, neuromesodermal (NM) stem cells give rise to both neural cells and paraxial presomitic mesoderm (PSM) cells, but what dictates PSM fate? Here, Terry Yamaguchi and colleagues show that a single transcription factor – mesogenin 1 (Msgn1) – acts as a master regulator of PSM development (p. 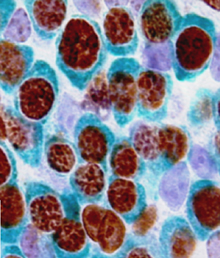 Embryonic germ cells display strikingly different fates with regard to mitosis and meiosis, depending on their sex. In female mice, germ cells switch from mitosis to meiosis shortly after reaching the foetal gonad where they generate the lifelong pool of oocytes. However, in males, meiosis and mitosis are actively repressed, and germ cells remain quiescent in the gonad until birth, when they resume mitosis and start generating spermatocytes. Here (p.
Embryonic germ cells display strikingly different fates with regard to mitosis and meiosis, depending on their sex. In female mice, germ cells switch from mitosis to meiosis shortly after reaching the foetal gonad where they generate the lifelong pool of oocytes. However, in males, meiosis and mitosis are actively repressed, and germ cells remain quiescent in the gonad until birth, when they resume mitosis and start generating spermatocytes. Here (p. 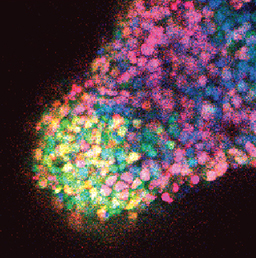 One of the first patterning events of embryogenesis occurs during gastrulation: three-dimensional (3D) cell movements reorganise the embryo, a mass of morphologically similar cells, into an axially organised structure with three germinal layers (endoderm, mesoderm and ectoderm). To date, two-dimensional (2D) culture models have failed to recapitulate such complex cell behaviours linking cell movement to cell fate. Here (p.
One of the first patterning events of embryogenesis occurs during gastrulation: three-dimensional (3D) cell movements reorganise the embryo, a mass of morphologically similar cells, into an axially organised structure with three germinal layers (endoderm, mesoderm and ectoderm). To date, two-dimensional (2D) culture models have failed to recapitulate such complex cell behaviours linking cell movement to cell fate. Here (p. 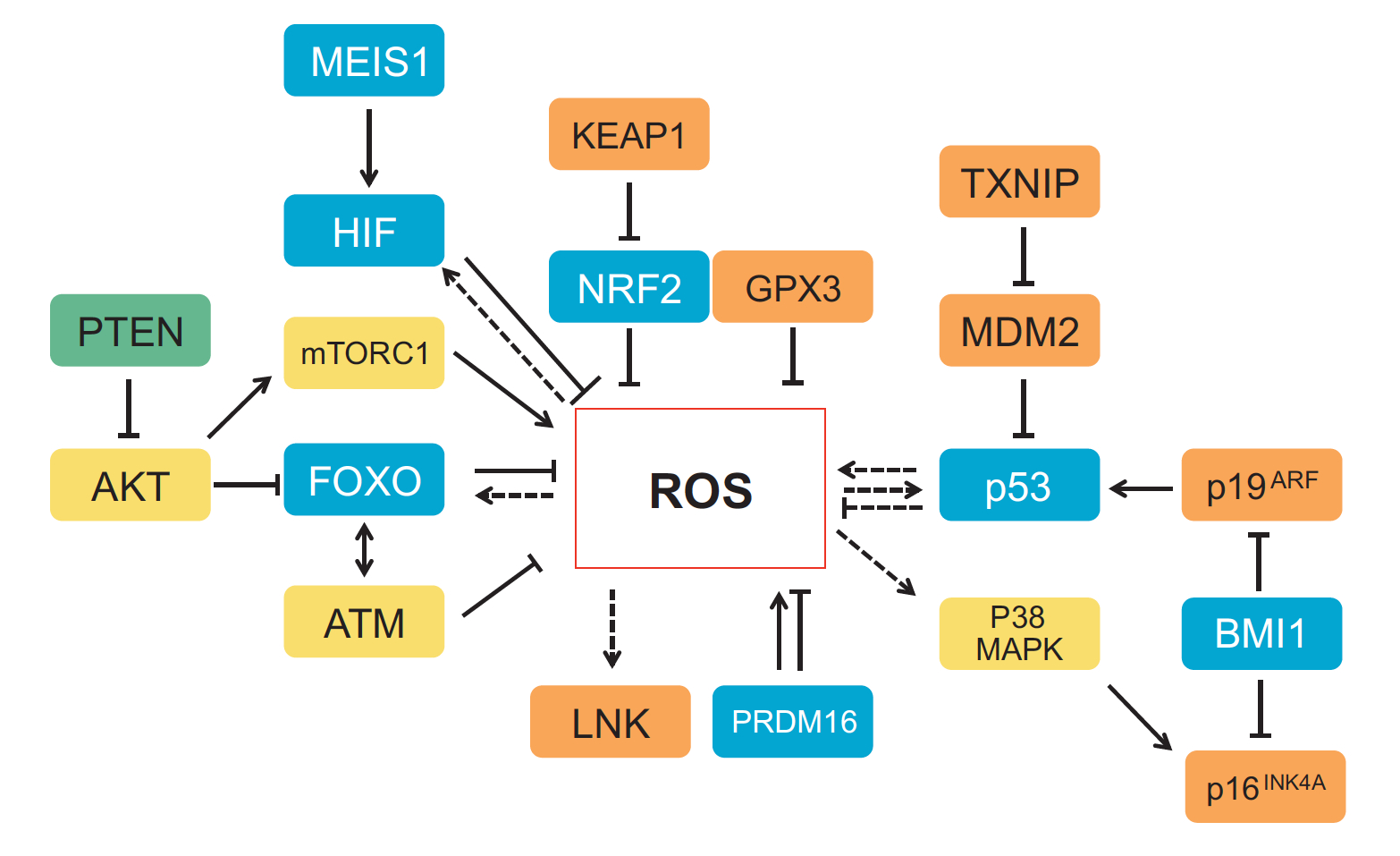 Recent work suggests that reactive oxygen species (ROS) can influence stem cell homeostasis and lineage commitment. In this Primer, Ghaffari and colleagues provide an overview of ROS signalling and its impact on stem cells, reprogramming and ageing. See the Primer on p.
Recent work suggests that reactive oxygen species (ROS) can influence stem cell homeostasis and lineage commitment. In this Primer, Ghaffari and colleagues provide an overview of ROS signalling and its impact on stem cells, reprogramming and ageing. See the Primer on p. 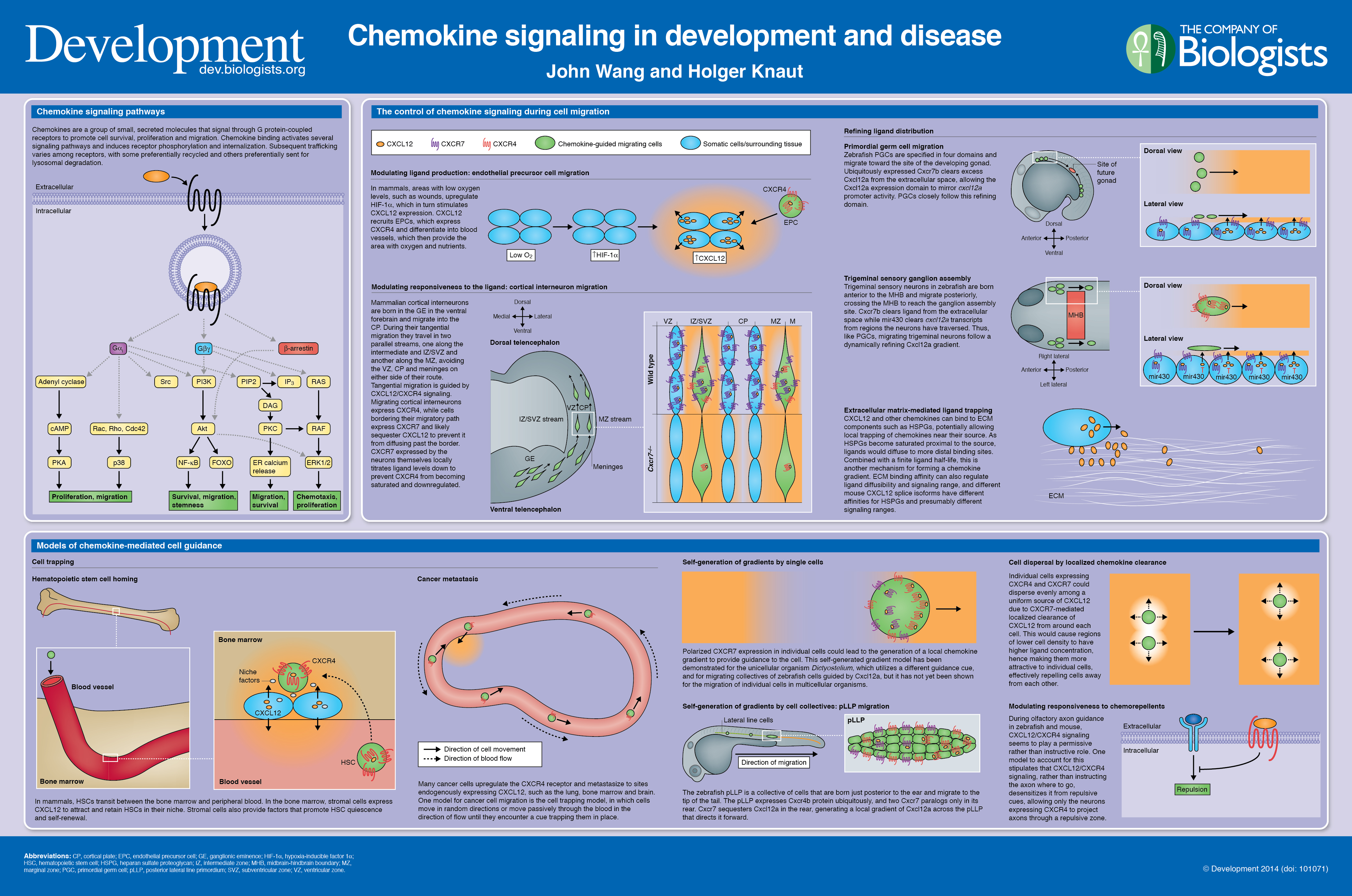 In our latest poster and companion article, Wang and Knaut provide an overview of chemokine signalling and some the chemokine-dependent strategies used to guide migrating cells. See the poster on p.
In our latest poster and companion article, Wang and Knaut provide an overview of chemokine signalling and some the chemokine-dependent strategies used to guide migrating cells. See the poster on p.  The development of plant leaves follows a common basic program, which can be modulated to generate a diverse range of leaf forms. Bar and Ori review recent work examining how plant hormones, transcription factors and tissue mechanics influence leaf development. See the Review on p.
The development of plant leaves follows a common basic program, which can be modulated to generate a diverse range of leaf forms. Bar and Ori review recent work examining how plant hormones, transcription factors and tissue mechanics influence leaf development. See the Review on p.  (No Ratings Yet)
(No Ratings Yet)











 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series