An interview with Christopher Wylie and Janet Heasman
Posted by the Node, on 18 November 2014
This interview first appeared in Development.
The 2014 Society for Developmental Biology (SDB) Lifetime Achievement Award was jointly awarded to Christopher Wylie and Janet Heasman in recognition of their outstanding and sustained contributions to the field. At the 73rd Annual SDB meeting, where they were presented with the award, we asked Chris and Janet about their careers and their advice for young researchers.
You both started off in medical school and then switched to a career in basic research. What inspired you to make the switch?
C: Actually,much of it was due to one of my professors, J. Z.Young, who did several things that enhanced basic science in medicine. Because students enter medical school directly from high school in England, they do not get exposed to research. J. Z. introduced a BSc-level science degree into the medical school curriculum. This was offered to students who did well in their medical examinations, and also served as entry into the MD/PhD program. Both Janet and I went with this route and just fell in love with biomedical research. A major part of the BSc course was a research project. I did mine with Ruth Bellairs and became fascinated by embryo development. I tried to make a movie of one of the experiments that we were doing but, in those days, making movies of embryos was a very complicated business. The movie (of a chick embryo) I was making went on all night, so I had to sit in the hot room for hours (there were no heated slide chambers in those days). Because of the heat, the grease in the microscope would melt and the body of the microscope would slowly slip down the column, so it had to be refocused after each image. By about 3 o’clock in the morning I was virtually naked and dripping with sweat! But I couldn’t believe my eyes as more and more embryonic structures appeared, as if by magic. Of course I had seen diagrams of development in my lectures, but I had never actually seen it happening. I was absolutely gobsmacked! That was my epiphany. And so I finished my BSc and was lucky enough to get offered a PhD place. Of course, the original objective was to then return to clinical medicine but I never did. I was offered a lectureship at University College, London, and that was the end of my clinical career.
J: I even did a year of clinical training (in those days medical school was two years of preclinical training and then three years of clinical school) but I didn’t enjoy it. I had the chance to go to Dartmouth College and work as a teaching assistant with Chris in developmental biology. While I was there I gave three lectures and I remember thinking to myself, if I can do this – if I can teach developmental biology successfully – then I am going to go back to England, pull out of my medical career and register as a PhD student. And that is what I did.
You’ve both been involved in setting up developmental biology institutes – in Cambridge, Minnesota and Cincinnati – and these are all continuing to flourish. What do you think is the key to success in a good research department or institute?
C: I think you have to have the right people…and money! If you recruit scientists who are willing to interact with each other and not compete, then you’ll probably have a successful institute, especially if it is well funded. Another thing that might seem trivial but proved to be very important was the tea room! It offered a centre where principals, group leaders, graduate students, postdocs and technicians could all meet and talk informally. Every morning at 10.30 at the Wellcome/CRC Institute in Cambridge (now the Gurdon Institute) we would all go up to the tea room for tea/coffee and chat. The things that we chatted about were not just ‘do you have an antibody against x that we can borrow?’, it was also ‘how do you think the institute could work better?’, ‘how can we improve the vivarium?’, ‘how is our financial situation?’. This day-by-day interaction was amazingly helpful for all aspects of the institute’s activities, not just our own research. It certainly made faculty meetings shorter because we had already discussed pretty well everything that came up.
J: Yes, the tea room worked really well in England.We tried very hard to make it work in the USA, but it was hard to persuade people to go to the tea room. No one would allow themselves enough time off to interact over tea! I also agree that the success of an institute depends on having the right people and being very careful in the job searches. It is absolutely essential to hire people who are interactive and who are going to collaborate with each other. It’s also important to have freedom – the freedom to be able to do science the way you want.
C: Yes, in Cambridge, the institute was funded by the Wellcome Trust and the Cancer Research Campaign, and they gave us a free hand. So we were largely buffered against Cambridge University politics and departmental structure. Similarly, in Minnesota I was just given the money and told ‘we need a centre for developmental biology’. And in Cincinnati I was given an even larger amount of money and told ‘build us a centre for developmental biology, Chris’. When you have such a financial advantage, it is not that difficult.
Chris, you have obviously been heavily involved with Development: you were Editor-in-Chief for a very long time and you made some pretty drastic changes to the journal when you started. How did you get involved with the journal and what prompted you to make those bold changes?
C: It all began when I was the Publications Officer of the British Society for Developmental Biology (BSDB). My job was to think up suitable symposium topics and identify people who could put a symposium together, edit the volume and to see it through to publication. At first, the papers presented at these symposia were published by Cambridge University Press as books, but the publication times were long and the costs were very high (and the profits to the society correspondingly small). So the BSDB moved to publishing their symposia as supplement volumes to The Journal of Embryology and Experimental Morphology (JEEM), which was owned and published by The Company of Biologists. The symposia attracted outstanding speakers (and therefore authors) from all over the world, and I couldn’t help noticing that the articles in the symposium supplements were generally more exciting than those in the parent journal. I thought, we are missing a lot of stuff in our journals in the UK. We need a journal that will capture the most recent advances in cellular, molecular and genetic approaches to developmental biology. So I went to several publishers and I said that I think there should be a journal (I had already called it Development in my mind) that captures all this stuff that the current British journals are missing. Lots of publishers took an interest in this. I was then approached by The Company of Biologists (who had heard about these discussions) and asked if I would take over the editorship of JEEM. I told them that I didn’t want to be the editor of JEEM but that I would consider being the editor of a different journal, one that had all the things that I thought should go into a topical developmental biology journal, and with a more modern format. Eventually they agreed. They said ‘Okay, do whatever you want.We will start a new journal, we will replace JEEM with this journal, and you can make whatever changes you want!’. And so I had a journal – Development – on my hands. It was absolute mayhem initially – it was probably June when we started and we had to have the issue out by January the following year – but, mercifully, the world’s developmental biologists reacted magnificently. I guess everyone else thought that there should be a journal like that, too.
That was over 25 years ago. How do you think the field has changed since then?
C: Well, I think the science – especially in terms of techniques and approaches – is changing all the time. In just one year, since I’ve retired, I can see by coming back to this meeting that there are techniques that we weren’t using just last year when I was in the lab.
J: Another big change is that everything is now online. The fact that no one uses libraries anymore has changed publishing, and it has changed interactions between people and groups.
C: It also means that, to some extent, it doesn’t really matter anymore where you publish, because people don’t go to libraries and open journals and go through them. They use searches and keywords so it doesn’t really matter whether your paper is published in what was formerly regarded as a ‘second tier’ journal or a premier journal; if your paper is good, it is going to be read.
J: And it is going to be reviewed in a very informal sense too – online. There are big pluses to that but there are also some minuses because sometimes only the people with the loudest ‘voices’ will be heard. Distinguishing gossip from good science is becoming more and more difficult these days.
C: Another thing that has changed a lot is the number of model organisms studied. When I was a student there were at least a dozen experimental model organisms being used to study development.We needed to learn their anatomy and embryogenesis to keep up with the genetic, molecular and cellular data coming out. However, as both the time and money required to set up the appropriate protocols and reagents became so great, the number of model organisms used contracted dramatically. So, during the time that I was an editor at Development,we ended up going from a dozen or so model organisms that made regular appearances in its pages to just four or five.
J: But now it has expanded again, which is really exciting. Techniques such as CRISPR have allowed us to go the other way. It was very noticeable at this meeting that there are again a variety of model organisms being used. Evolution is at the centre again.
Science seems to be quite competitive these days, and it is getting increasingly difficult to get an academic tenure track position. What is your advice to young people who are starting out?
J: To recognise that it is very important to develop your own ideas of what research you want to do, even as a graduate student; don’t wait until the postdoc stage. Actively think about where you want your career to go and look for the labs that will fit what you want to do, rather than looking for the lab that will take you. You will be impressive in an interview if you know what you want to do. Rather than thinking ‘how am I going to finish my graduate studies?’, ‘how am I going to get my first fellowship?’ and so on, think ‘what am I really interested in answering?’, ‘what biological question do I want to answer?’ and ‘which model system should I use?’.
C: I think you have to choose the right postdoc position. This is really, really important. Also, you need to find good mentorship. That applies at all levels. If you don’t have a good PhD mentor, who will look after your next stage by being honest about labs that you should or should not go to as a postdoc, you are at a serious disadvantage. Similarly, you need a good postdoc mentor whowill advise you about which places you should think about going to for getting a job (and where you should not think about going to!). And then, when you are an assistant professor, you need really good mentorship for all aspects of your research life, including: research approaches, collaborations that will help you, grant applications, recruitment of lab personnel, disputes that occur in your lab, where you should send your papers, what committees you should (and shouldn’t) sit on.Young faculty are not ready for that kind of stuff, and they need senior faculty to act as good mentors and who can offer them advice, read papers and grants, and provide honest feedback.
You’ve both had very successful careers working as a husband-and-wife team while raising a family. What is your advice to younger people who are starting off in terms of work-life balance?
J: I think my advice is just to do it. There is no easy way. And that is true whether you are a scientist or working in banking or business. It is a balancing act that requires constant communication. I think scientists in some ways have it easy if they work together. I can always say I know where Chris is – he is in the lab! I know how to contact him there. I actually think that science is a good career for husband and wife teams. And don’t take yourself, and science, too seriously; at the end of the day you are a small cog in a big wheel and your kids carry on after you.
C: And be prepared to work shifts. We used to do that a lot.
J: And my advice to young women is: don’t put off having kids until the time is right. Because the time is never right.
C: The problem with putting off kids until the ‘right time’ is that you have to set criteria for the right time, and it is hard to know when these have been met. It is easy to say ‘we will have children when…’. But of course you never reach the ‘when’.
And, finally, what would people be surprised to find out about you?
C: I don’t think there is much about me that would surprise people. I think the basis for my success in editorship and leadership has been that I’m not a particularly complicated or devious person. So nobody ever needed to try to second guess me! I suppose that, given the public persona required to teach, lead a research centre, or edit a major journal, people might be surprised to know that I’mnot a gregarious person. The best part of my career has been working quietly in the lab, trying to discover things, reading about the discoveries others have made (I really miss the library), and trying to integrate it all into new knowledge.



 (6 votes)
(6 votes) Primordial germ cells (PGCs) are unipotent – they go on to form germline stem cells and gametes. However, they are believed to possess latent pluripotency, allowing them to produce the next generation during the normal life-cycle, and to be reprogrammed to pluripotent embryonic germ cells (EGCs) upon experimental manipulation. Various protocols for EGC establishment have been reported, with varying efficiency. Yasuhisa Matsui and colleagues now report a highly efficient method for mouse PGC-to-EGC conversion, using Akt activation in concert with bFGF and LIF treatment (p.
Primordial germ cells (PGCs) are unipotent – they go on to form germline stem cells and gametes. However, they are believed to possess latent pluripotency, allowing them to produce the next generation during the normal life-cycle, and to be reprogrammed to pluripotent embryonic germ cells (EGCs) upon experimental manipulation. Various protocols for EGC establishment have been reported, with varying efficiency. Yasuhisa Matsui and colleagues now report a highly efficient method for mouse PGC-to-EGC conversion, using Akt activation in concert with bFGF and LIF treatment (p.  The vasculature of the central nervous system (CNS) is highly specialised, characterised by the formation of the blood-brain barrier that prevents leakage of vascular contents into the brain. Various molecules and pathways have been implicated in regulating angiogenesis in the CNS, including the αVβ8 integrin and members of the TGFβ pathway. It is thought that αVβ8 integrin expressed in the neuroepithelium regulates TGFβ signalling in the endothelium. On p.
The vasculature of the central nervous system (CNS) is highly specialised, characterised by the formation of the blood-brain barrier that prevents leakage of vascular contents into the brain. Various molecules and pathways have been implicated in regulating angiogenesis in the CNS, including the αVβ8 integrin and members of the TGFβ pathway. It is thought that αVβ8 integrin expressed in the neuroepithelium regulates TGFβ signalling in the endothelium. On p. The liver possesses a remarkable capacity to regenerate, but what is the source of new cells during regeneration? The two epithelial cell populations – hepatocytes and cholangiocytes – can proliferate upon injury, but there is also evidence for the existence of an adult stem/progenitor cell, the liver progenitor cell (LPC), which resides in or near bile ducts and has both hepatocytic and cholangiocytic potential. Here (p.
The liver possesses a remarkable capacity to regenerate, but what is the source of new cells during regeneration? The two epithelial cell populations – hepatocytes and cholangiocytes – can proliferate upon injury, but there is also evidence for the existence of an adult stem/progenitor cell, the liver progenitor cell (LPC), which resides in or near bile ducts and has both hepatocytic and cholangiocytic potential. Here (p. During heart development, the coronary vasculature forms by establishment of an endothelial plexus that expands around the heart. Current evidence suggests that the sinus venosus (SV), endocardium and proepicardium may all contribute to coronary development. However, the relative contributions of these sources and the molecular mechanisms regulating coronary angiogenesis are still unclear. In a detailed lineage-tracing analysis in mouse (p.
During heart development, the coronary vasculature forms by establishment of an endothelial plexus that expands around the heart. Current evidence suggests that the sinus venosus (SV), endocardium and proepicardium may all contribute to coronary development. However, the relative contributions of these sources and the molecular mechanisms regulating coronary angiogenesis are still unclear. In a detailed lineage-tracing analysis in mouse (p.  The 2014 Society for Developmental Biology (SDB) Lifetime Achievement Award was jointly awarded to Christopher Wylie and Janet Heasman in recognition of their outstanding and sustained contributions to the field. At the 73rd Annual SDB meeting, where they were presented with the award, we asked Chris and Janet about their careers and their advice for young researchers. See the Spotlight on p.
The 2014 Society for Developmental Biology (SDB) Lifetime Achievement Award was jointly awarded to Christopher Wylie and Janet Heasman in recognition of their outstanding and sustained contributions to the field. At the 73rd Annual SDB meeting, where they were presented with the award, we asked Chris and Janet about their careers and their advice for young researchers. See the Spotlight on p. 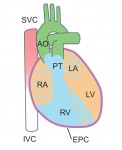 Kenneth Chien and colleagues discuss how insights into the molecular and cellular framework underlying cardiac development can be used to guide the
Kenneth Chien and colleagues discuss how insights into the molecular and cellular framework underlying cardiac development can be used to guide the 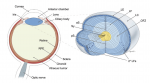 Although lens induction has been studied for over 100 years, recent findings have revealed a myriad of signaling pathways and gene regulatory networks that are required for lens formation in vertebrates. Ales Cvekl and Ruth Ashery-Padan summarize recent progress in the field, emphasizing the interplay between the diverse regulatory mechanisms employed to form lens progenitor and precursor cells. See the Review on p.
Although lens induction has been studied for over 100 years, recent findings have revealed a myriad of signaling pathways and gene regulatory networks that are required for lens formation in vertebrates. Ales Cvekl and Ruth Ashery-Padan summarize recent progress in the field, emphasizing the interplay between the diverse regulatory mechanisms employed to form lens progenitor and precursor cells. See the Review on p.  (No Ratings Yet)
(No Ratings Yet)