Hello! I’m Leila, a finishing PhD student in Patrícia Beldade’s lab at the Instituto Gulbenkian de Ciência, Portugal. We work on different topics within Evolutionary Developmental Biology, Evo-Devo, with the common interest on how development contributes to intra- and inter-specific variation and can influence evolutionary processes: developmental hierarchies (me), developmental plasticity, and the origin of novelty. The lab does not focus on a particular organism. Rather, we have flies, butterflies and ants. Because I’m interested in a highly diverse group – both taxonomically and morphologically – that can be manipulated experimentally, I chose butterflies.
Most of my work uses the African species Bicyclus anynana, established in the 80’s and living happily ever after in the lab. Butterflies are holometabolous insects, which means they go through four metamorphic stages: embryo (egg), larva (caterpillar), pupa (chrysalis), and adult (butterfly, Fig. 1A and 2A). For B. anynana, this cycle takes about 4 weeks at 27°C, and twice as much at 19°C. These are the temperatures that, in the lab, induce what are the natural wet- and dry-season phenotypes, respectively. Wet and dry morphs have different wing color patterns and life histories. The lab stock population has much genetic variation, which allows for artificial selection of distinct wing pattern and life history traits; they respond to artificial selection with particular ease. Many of our studies concentrate on a particular wing pattern element called the eyespot, which develops at the end of the larval stage and throughout the pupal stage. Eyespots are serially repeated structures, which are ideal for studies of modularity, and eyespots are also evolutionary novelties, that is, they only evolved within this group. And, of course, they are experimentally tractable: you can do many surgical manipulations and the prospective butterfly comes out just fine. I mean, fine – for us. This system is particularly interesting for understanding cell-fate determination because the wing is a 2D structure composed of parallel arrays of cells where each cell corresponds to one fate (or color, Fig. 2B). That added to the possibility of dissection of (much larger than flies) tissues, of tissue transplants, of pharmacological approaches by injection or tissue culture, of gene expression assays of any kind; genetic/genomic resources; and growing transgenic tools shines butterflies in the spotlight of Evo and Devo studies.
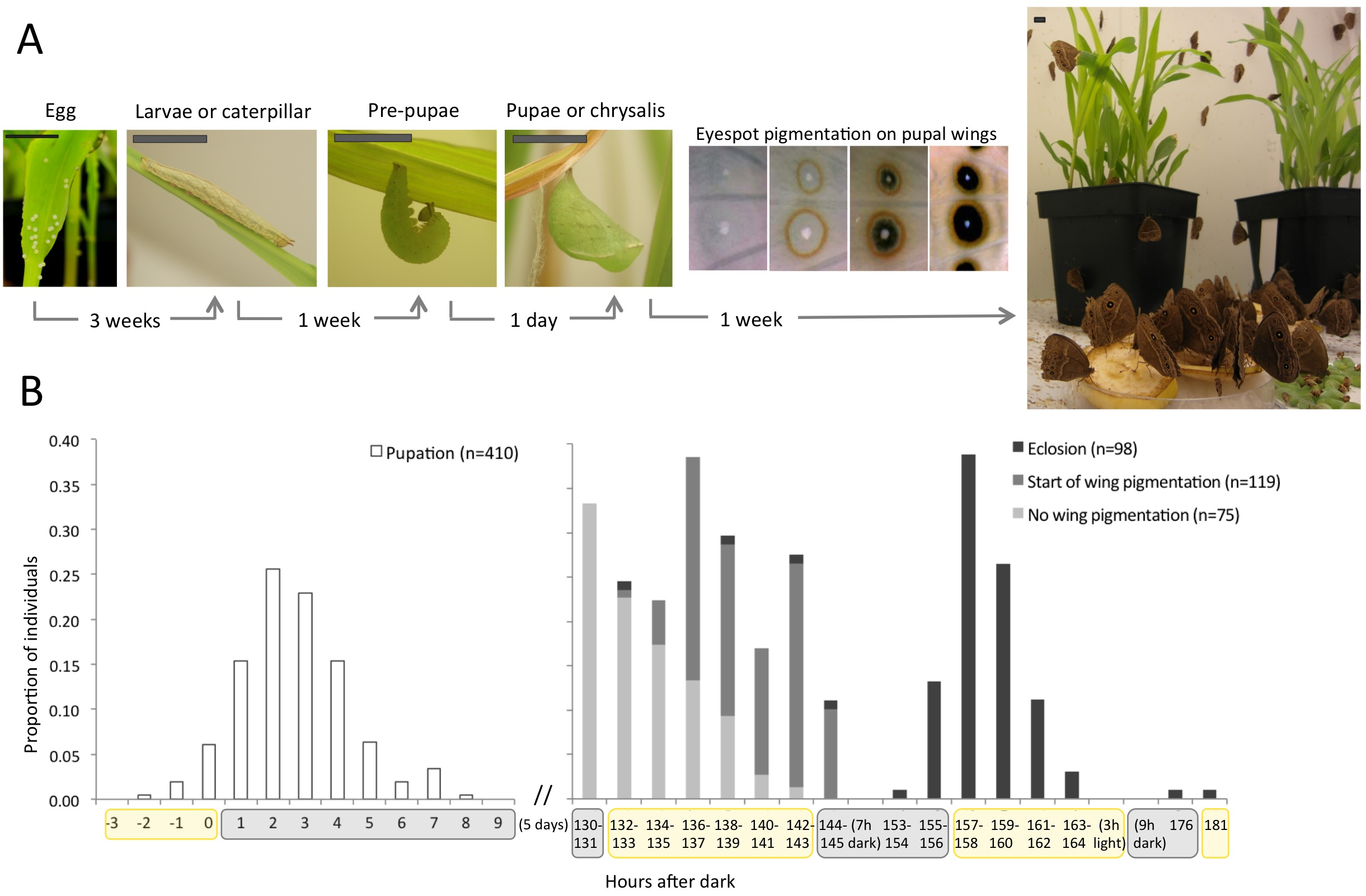
(A) Life cycle of butterflies, with time corresponding to Bicyclus anynana (Satyrinae, Nymphalidae) development at 27°C. At the end of the pupal stage, pigmentation takes place, here illustrated by the orderly pigment deposition on wings to form patterns elements called eyespots. Scale bar: 1cm. (B) Day-night cycles are associated to many life-history transitions including when pupation (left panel), the onset of wing pigmentation, and eclosion (right panel) occur. [click in the image to make it bigger]
Studying comparative development relies on having as many species in captivity as possible. Butterflies can be bred or purchased online (‘normal’ people do that for teaching life cycles in schools, or releasing them at weddings). Among species available for lab studies, we count on B. anynana (Fig. 1), buck-eye Junonia coenia (Fig. 2), Heliconius (beautiful example of mimmicry), speckled-wood Pararge aegeria, the cabbage butterfly Pieris rapae and P. brassicae, the migratory monarch Danaus plexippus, and Vanessa cardui (feeds on nettle – really painful working with these). You cannot, however, have butterflies at any time because many species hibernate or are univoltine, i.e., one generation per year. But whenever spring comes, it is time to get your net (and camera; it is memorable) or set up your trap. If you try doing this yourself, you will probably catch a species that needs real sunlight to grow, or doesn’t like to be stared at while doing, you know, reproduction, or or or. Even though many species can be bred, it’s not easy. Better going to butterfly houses, where all the laborious work is done with a smile in their faces.
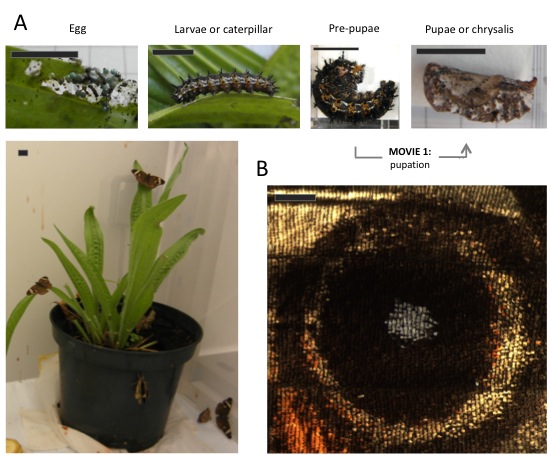
(A) Life cycle of Junonia coenia (Nymphalinae, Nymphalidae): Movie 1 shows the transition from pre-pupa to pupa. Scale bar: 1cm. (B) Butterfly wings are 2D structures composed by juxtaposition of cells in parallel rows as tiles in a roof, and each cell bears a single color. The image shows an eyespot that, within this arrangement, forms concentric rings of different colors. Scale bar: 1mm.
Real-time recording of pupation in Junonia coenia. The prospective pupa strips the black larval epidermis by whole-body contractions (refer to Fig. 2 for before-and-after stages). At the end of this movie, the location of eyes, proboscis, antennae, and wings can be seen given the cuticle is much thinner in the boundaries between organs. These “pre-cuts” help the eclosing butterfly to break the pupal cage.
Food and hygiene are critical aspects of animal breeding, as any developmental biologist knows. Since a lab usually needs food in almost-industrial scale, one either has to use artificial diets or cultivate crops. Our species is a grass-eater and we feed larvae on maize, such that the lab weekly rotates in agricultural, maize sowing, tasks. We can get seeds from popcorn or from local cooperatives, but never transgenic, engineered to resist “pests,” like caterpillars. In fact, our maize greenhouse, a warm and moist environment with endless food and no predators, is a dreamland for butterflies and many other arthropods. We often need to release spiders, aphids, other Lepidopterans (moths love our greenhouse), the vast world of Dipterans et al.; but also charming vertebrates such as our resident gecko. For hygiene, we constantly bleach eggs and the butterfly incubators (a controlled environment, with authorized Metazoans only), daily clean and spray cages with hospital sterilizing agents, change gloves between cages, have all materials washed and bleached, and so on. Even with all this care, diseases can spread quickly and once, no matter what we did, the poor fellows were getting sicker and sicker. The entire lab mobilized for a day of master cleaning. Picture this: dozens of butterfly cages stacked like apartments, a group of very mature scientists with labcoats, masks, gloves; sweeping, layering the butterfly facility with detergent-water-bleach-water, UV lamps, under the misty atmosphere of dust dancing along “I will survive” for so long it made us dizzy. There is no way that wouldn’t form a deep bond between us, so we repeat the ritual every semester.
A typical day in a butterfly lab involves feeding larvae – they walk to the new leaves so we only need disposing old “deciduous” maize pots; cleaning their cages; giving adults their banana; freezing eclosed adults from an experiment, and all trash to make sure nothing stays alive; collecting, bleaching, and counting eggs to establish a new generation; and finding green pre-pupae camouflaged in green leaves for experiments of the next day or week. This usually takes about a third of a day, so with the remaining time we do wet-lab and office duties. Similar to what Andrew Mathewson said in “A day in the life of a zebrafish lab,” butterflies are somehow in the middle of the frenetic rhythm of yeast, worms, and flies but not so long as mice and Arabidopsis. So usually we run a couple experiments in parallel and it’s not uncommon to start the day in the tropical 27°C incubator, get timed pupae and start running gene expression protocols, proceed to the dark and cold microscopy room for immunohistochemistries that finished, perform wing transplants or DNA/RNA extraction or set up assays in tissue culture, return to the incubator and turn on the camera to record pupation time.
As I follow the sequential, hierarchical stages of development, I keep close track of (their) time, which often compromises the notion of weekday and weekend. We take time-lapsed photographs during the night to know very exactly when pupation occurred. The pupal stage follows circadian cues (Fig. 1B). When final instar larvae are done eating, they crawl into a hidden place during the night, curl up and get immobilized in the pre-pupal stage, when they reorganize their innards. One day later, shortly after lights go off, they pupate (Movie 1). Five days pass and pigmentation begins in their eyes (Movie 2), wings, antennae, legs, and whole body. To characterize the progression of pigmentation, I dissect late pupae for every single of the last 48h of their development. It is great to, as a job, study how butterfly wings develop and get their colors.
Pigmentation in the eyes is already visible through the pupal cage in fifth-day pupae of B. anynana. Movie assembled from time-lapse images taken every 5min during 7h.
The same individual of Movie 2 in its last day of pupal life, when all organs are ready and final sclerotization takes place; sclerotization is the process by which cuticular cells harden, rendering them impermeable. Movie assembled from time-lapse images taken every 5min during 6h.
Pigmentation starts in the afternoon of the 5th day and colors on eyes and wings are already visible through the pupal cage in the morning of the 6th day. Next day, 2h after light goes on, the eclosing butterfly breaks the softened pupal cage (Movie 3). Wings go first, then head, then abdomen; their long tongue, or proboscis, curls (a synapomorphy!); their wings stretch and pump haemolymph so they expand to become 2x, 3x, 4x larger and finally, an hour later, they attempt their first flight – and usually fall. Many important steps happen in the dark, probably a protective strategy for sessile pupae to move when no bird sees. Also, as butterflies are sensitive to temperature, it makes sense to be ready to fly with the rising sun, find nectar, find mates, and fill the world with joy.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
 (10 votes)
(10 votes)
 Loading...
Loading...


 (6 votes)
(6 votes)

 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series  (No Ratings Yet)
(No Ratings Yet)
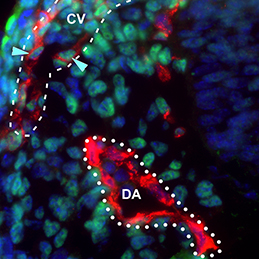 The dorsal aorta (DA) and the cardinal vein (CV) are the first vascular pair to form during development. Although it is generally accepted that the DA derives from angioblasts, the cellular origin of the mammalian CV is currently unknown. Now, on
The dorsal aorta (DA) and the cardinal vein (CV) are the first vascular pair to form during development. Although it is generally accepted that the DA derives from angioblasts, the cellular origin of the mammalian CV is currently unknown. Now, on 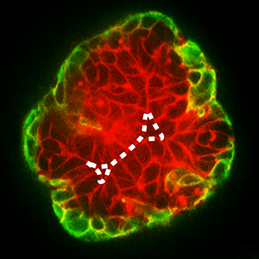 During development, select epithelial cells must undergo asymmetric division in order to generate a stratified epithelium. Previous studies have shown that basally positioned epithelial stem cells orchestrate this process, but the mammary epithelium has two distinct cell layers inside the basement membrane, so it remains unclear which cell type is responsible for stratification. Using elegant time-lapse imaging of three-dimensional mouse mammary epithelial cultures, Andrew Ewald and colleagues now reveal (
During development, select epithelial cells must undergo asymmetric division in order to generate a stratified epithelium. Previous studies have shown that basally positioned epithelial stem cells orchestrate this process, but the mammary epithelium has two distinct cell layers inside the basement membrane, so it remains unclear which cell type is responsible for stratification. Using elegant time-lapse imaging of three-dimensional mouse mammary epithelial cultures, Andrew Ewald and colleagues now reveal (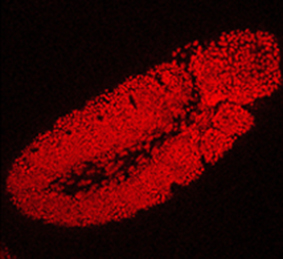 The Drosophila Trithorax (TRX) protein plays a key role in maintaining active transcription of many master cell fate regulatory genes. Previously, TRX was thought to function mainly at the promoter by depositing the histone H3K4 trimethylation mark (H3K4me3) via its catalytic SET domain. However, recent findings have challenged this view, prompting new investigations into the function of TRX. Now, on
The Drosophila Trithorax (TRX) protein plays a key role in maintaining active transcription of many master cell fate regulatory genes. Previously, TRX was thought to function mainly at the promoter by depositing the histone H3K4 trimethylation mark (H3K4me3) via its catalytic SET domain. However, recent findings have challenged this view, prompting new investigations into the function of TRX. Now, on 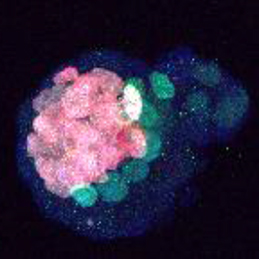 The transcription factor Oct4 is well known for its role in maintaining pluripotency in vitro and for preventing ectopic differentiation of early embryos in vivo. Recent evidence also suggests a role for Oct4 in lineage specification; however, little is known about how this occurs and the manner in which Oct4 is required. Now, on
The transcription factor Oct4 is well known for its role in maintaining pluripotency in vitro and for preventing ectopic differentiation of early embryos in vivo. Recent evidence also suggests a role for Oct4 in lineage specification; however, little is known about how this occurs and the manner in which Oct4 is required. Now, on 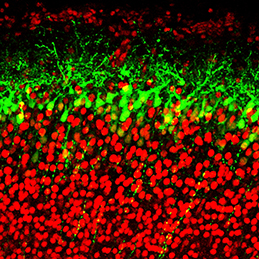 The generation of layer-specific cortical neurons is fundamental to the circuitry of the developing and adult brain. Although the intrinsic drivers of neuronal specification are becoming increasingly understood, the extrinsic signals that guide migration and consolidate post-mitotic neuronal identity are less clear. In this issue (
The generation of layer-specific cortical neurons is fundamental to the circuitry of the developing and adult brain. Although the intrinsic drivers of neuronal specification are becoming increasingly understood, the extrinsic signals that guide migration and consolidate post-mitotic neuronal identity are less clear. In this issue (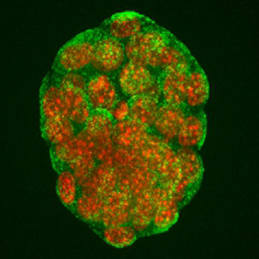 The production of healthy offspring depends on many factors spanning from intrinsic genetic elements to variations in the in uteroenvironment. Poor maternal diet is associated with increased risk of cardiovascular, metabolic and behavioural disorders during later life of the offspring, but how the developing embryo copes with maternal dietary stress has not been well characterised. Now, on
The production of healthy offspring depends on many factors spanning from intrinsic genetic elements to variations in the in uteroenvironment. Poor maternal diet is associated with increased risk of cardiovascular, metabolic and behavioural disorders during later life of the offspring, but how the developing embryo copes with maternal dietary stress has not been well characterised. Now, on  It has been known for years, based on studies of Drosophila, that viable cells can be eliminated by their neighbours through a process termed cell competition. New studies in mammals have revealed that this process is universal and that many factors and mechanisms are conserved. Here, Amoyel and Bach provide an overview of the mechanistic steps involved in cell competition and discuss recent advances in the field, which have shed light on how and why cell competition exists in developing and adult organisms. See the Review on p.
It has been known for years, based on studies of Drosophila, that viable cells can be eliminated by their neighbours through a process termed cell competition. New studies in mammals have revealed that this process is universal and that many factors and mechanisms are conserved. Here, Amoyel and Bach provide an overview of the mechanistic steps involved in cell competition and discuss recent advances in the field, which have shed light on how and why cell competition exists in developing and adult organisms. See the Review on p. 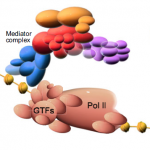 Mediator is a multiprotein complex that is required for gene transcription by RNA polymerase II. Here, Yin and Wang describe the most recent advances in understanding the mechanisms of Mediator function, with an emphasis on its role during development and disease. See the Primer on p.
Mediator is a multiprotein complex that is required for gene transcription by RNA polymerase II. Here, Yin and Wang describe the most recent advances in understanding the mechanisms of Mediator function, with an emphasis on its role during development and disease. See the Primer on p.