Here are the highlights from the current issue of Development:
In-cyst-ing on germ cell development
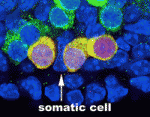 During gametogenesis in many organisms, germ cells undergo synchronous, incomplete divisions just before meiosis that generate interconnected cell groups known as cysts, but does a cyst stage always occur during mammalian gametogenesis? Here (p. 2075), by lineage marking the progeny of primordial germ cells (PGCs) in embryonic mouse gonads, Lei Lei and Allan Spradling show that mouse PGCs initially develop into cysts containing two, four or eight cells. In female gonads, these cysts fragment into smaller cysts that associate with the cysts of unrelated progenitors. Interestingly, the number of female cysts per PGC at the time of meiotic entry corresponds closely with the number of primordial follicles produced by each progenitor, which suggests that cyst fragmentation determines the number of oocytes and possibly also their quality. Male cyst cells also break apart, the researchers report, which amplifies the number of spermatogonial stem cells. Together, these results indicate that cyst formation and cyst fragmentation are fundamental stages in the development of male and female mouse gametes.
During gametogenesis in many organisms, germ cells undergo synchronous, incomplete divisions just before meiosis that generate interconnected cell groups known as cysts, but does a cyst stage always occur during mammalian gametogenesis? Here (p. 2075), by lineage marking the progeny of primordial germ cells (PGCs) in embryonic mouse gonads, Lei Lei and Allan Spradling show that mouse PGCs initially develop into cysts containing two, four or eight cells. In female gonads, these cysts fragment into smaller cysts that associate with the cysts of unrelated progenitors. Interestingly, the number of female cysts per PGC at the time of meiotic entry corresponds closely with the number of primordial follicles produced by each progenitor, which suggests that cyst fragmentation determines the number of oocytes and possibly also their quality. Male cyst cells also break apart, the researchers report, which amplifies the number of spermatogonial stem cells. Together, these results indicate that cyst formation and cyst fragmentation are fundamental stages in the development of male and female mouse gametes.
Meiotic progression, ETC
 During mitosis, ubiquitylation of the anaphase inhibitor securin by the E3 ligase anaphase-promoting complex/cyclosome (APC/C) targets securin for proteolysis, which triggers sister chromatid separation. Securin is also implicated in meiotic progression. Thus, during both mitosis and meiosis, securin expression must be tightly regulated to enable its timely action. On p. 2149, Risa Kitagawa and co-workers investigate the mechanism underlying this regulation by analysing the subcellular localisation of the securin IFY-1 during C. elegans development. IFY-1 expression is high in the cytoplasm of germ cells, they report, but declines following meiosis I, and remains low during meiosis II and following mitoses. The researchers identify the HECT-E3 ligase ETC-1 as a regulator of the cytoplasmic levels of IFY-1 and CYB-1 (cyclin B1), and show that ETC-1 ubiquitylates IFY-1 and CYB-1 in vitro in the presence of the E2 enzyme UBC-18. These and other data suggest that a novel mechanism, mediated by ETC-1, cooperates with APC/C to regulate the timing of meiosis during germ cell development in C. elegans.
During mitosis, ubiquitylation of the anaphase inhibitor securin by the E3 ligase anaphase-promoting complex/cyclosome (APC/C) targets securin for proteolysis, which triggers sister chromatid separation. Securin is also implicated in meiotic progression. Thus, during both mitosis and meiosis, securin expression must be tightly regulated to enable its timely action. On p. 2149, Risa Kitagawa and co-workers investigate the mechanism underlying this regulation by analysing the subcellular localisation of the securin IFY-1 during C. elegans development. IFY-1 expression is high in the cytoplasm of germ cells, they report, but declines following meiosis I, and remains low during meiosis II and following mitoses. The researchers identify the HECT-E3 ligase ETC-1 as a regulator of the cytoplasmic levels of IFY-1 and CYB-1 (cyclin B1), and show that ETC-1 ubiquitylates IFY-1 and CYB-1 in vitro in the presence of the E2 enzyme UBC-18. These and other data suggest that a novel mechanism, mediated by ETC-1, cooperates with APC/C to regulate the timing of meiosis during germ cell development in C. elegans.
Tentacles illuminate appendage evolution
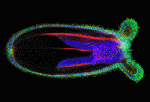 Evolution of the capacity to form appendages (secondary outgrowths from the principal embryonic axes) potentiated the diversification of animal body plans. The basic mechanisms that underlie appendage growth in bilaterian model systems have been identified but little is known about appendage development in pre-bilaterians. Now, on p. 2212, Matthew Gibson and colleagues report that three processes contribute to tentacle development in the sea anemone Nematostella vectensis. The initial step in tentacle development in this early-branching metazoan, they report, is the formation of a thickened ectodermal placode at the oral pole that progressively divides into four distinct tentacle domains, probably through the restricted expression of key effector genes. Next, Notch signalling triggers apicobasal thinning of the tentacular ectoderm, which drives the elongation of these primordia. Concomitantly, oriented cell rearrangements help to shape the elongating tentacles. By defining the mechanism of embryonic appendage development in an early-branching metazoan, these findings provide a foundation for understanding the evolutionary diversification of animal body plans.
Evolution of the capacity to form appendages (secondary outgrowths from the principal embryonic axes) potentiated the diversification of animal body plans. The basic mechanisms that underlie appendage growth in bilaterian model systems have been identified but little is known about appendage development in pre-bilaterians. Now, on p. 2212, Matthew Gibson and colleagues report that three processes contribute to tentacle development in the sea anemone Nematostella vectensis. The initial step in tentacle development in this early-branching metazoan, they report, is the formation of a thickened ectodermal placode at the oral pole that progressively divides into four distinct tentacle domains, probably through the restricted expression of key effector genes. Next, Notch signalling triggers apicobasal thinning of the tentacular ectoderm, which drives the elongation of these primordia. Concomitantly, oriented cell rearrangements help to shape the elongating tentacles. By defining the mechanism of embryonic appendage development in an early-branching metazoan, these findings provide a foundation for understanding the evolutionary diversification of animal body plans.
Ephrin B1 sticks up for developing brain
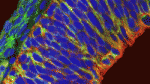 The apical membrane of apical progenitors (APs) in the developing mammalian cortex is involved in cell-cell and cell-extracellular matrix (ECM) adhesion events that maintain the integrity of the developing neuroepithelium. Apical adhesion is important for several aspects of neural development but its regulation is poorly understood. Here (p. 2082), Alice Davy and colleagues investigate the function of ephrin B1, a cell-surface protein that activates cellular signalling on binding ephrin receptors (via reverse signalling), in the morphogenesis of the developing mouse cortex. The researchers report that some Efnb1-deficient embryos display exencephaly and exhibit morphological alterations of the neuroepithelium that suggest defects in neural tube closure. Ephrin B1 is required in APs to maintain apical adhesion, they report, and ephrin B1 reverse signalling controls integrin-based cell-ECM apical adhesion through inhibition of ADP-ribosylation factor 6. These results suggest that ephrin B1 maintains the apical adhesion of APs during cortical development, a function that is essential for neural tube morphogenesis.
The apical membrane of apical progenitors (APs) in the developing mammalian cortex is involved in cell-cell and cell-extracellular matrix (ECM) adhesion events that maintain the integrity of the developing neuroepithelium. Apical adhesion is important for several aspects of neural development but its regulation is poorly understood. Here (p. 2082), Alice Davy and colleagues investigate the function of ephrin B1, a cell-surface protein that activates cellular signalling on binding ephrin receptors (via reverse signalling), in the morphogenesis of the developing mouse cortex. The researchers report that some Efnb1-deficient embryos display exencephaly and exhibit morphological alterations of the neuroepithelium that suggest defects in neural tube closure. Ephrin B1 is required in APs to maintain apical adhesion, they report, and ephrin B1 reverse signalling controls integrin-based cell-ECM apical adhesion through inhibition of ADP-ribosylation factor 6. These results suggest that ephrin B1 maintains the apical adhesion of APs during cortical development, a function that is essential for neural tube morphogenesis.
Fascin-ating melanoblast migration
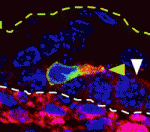 The actin-bundling protein fascin 1 is expressed at specific times and places during development, and is often expressed in tumours. Fascin 1 has a clear role in cell migration in vitro but what is its role in vivo? To answer this question, Laura Machesky and co-workers have been studying the role of fascin 1 in the mouse melanocyte lineage and in human melanoma cells (p. 2203). They report that fascin 1 knockout mice have hypopigmentation defects that arise from reduced melanoblast migration and proliferation during embryogenesis. By studying live embryo skin explants, they show that fascin 1-null melanoblasts migrate and differentiate in the skin but with a lower efficiency than wild-type melanoblasts. They also show that fascin 1 depletion slows melanoblast proliferation in vivo and human melanoma cell growth in vitro. The researchers suggest, therefore, that transient expression of fascin 1 in mouse melanoblasts during embryogenesis promotes their migration and proliferation, and that fascin 1 expression may aid the proliferation and invasion of some tumour cells.
The actin-bundling protein fascin 1 is expressed at specific times and places during development, and is often expressed in tumours. Fascin 1 has a clear role in cell migration in vitro but what is its role in vivo? To answer this question, Laura Machesky and co-workers have been studying the role of fascin 1 in the mouse melanocyte lineage and in human melanoma cells (p. 2203). They report that fascin 1 knockout mice have hypopigmentation defects that arise from reduced melanoblast migration and proliferation during embryogenesis. By studying live embryo skin explants, they show that fascin 1-null melanoblasts migrate and differentiate in the skin but with a lower efficiency than wild-type melanoblasts. They also show that fascin 1 depletion slows melanoblast proliferation in vivo and human melanoma cell growth in vitro. The researchers suggest, therefore, that transient expression of fascin 1 in mouse melanoblasts during embryogenesis promotes their migration and proliferation, and that fascin 1 expression may aid the proliferation and invasion of some tumour cells.
Plant vascular tissue lines up
 Plant vascular tissue is derived from the cambium and procambium, which are meristems that contain long thin stem cells that divide down their long axis. These highly organised divisions produce characteristic files of cells in the xylem and ensure spatial separation of the xylem and phloem. The ligand-receptor pair CLE41-PXY regulates vascular cell division, vascular organisation and xylem differentiation. Now, on p. 2224, Simon Turner and colleagues provide new insights into the factors that act downstream of PXY in Arabidopsis. The researchers show that WOX4, which encodes a transcription factor previously shown to act downstream of PXY, acts redundantly with WOX14 to regulate vascular cell division but that neither gene regulates vascular organisation. Moreover, they identify an interaction between PXY and the receptor kinase ERECTA that affects the organisation of the vascular tissue but not its rate of cell division. The researchers propose, therefore, that PXY and ERECTA signalling integrate cell division and vascular organisation in the Arabidopsis stem, thereby ensuring normal stem formation.
Plant vascular tissue is derived from the cambium and procambium, which are meristems that contain long thin stem cells that divide down their long axis. These highly organised divisions produce characteristic files of cells in the xylem and ensure spatial separation of the xylem and phloem. The ligand-receptor pair CLE41-PXY regulates vascular cell division, vascular organisation and xylem differentiation. Now, on p. 2224, Simon Turner and colleagues provide new insights into the factors that act downstream of PXY in Arabidopsis. The researchers show that WOX4, which encodes a transcription factor previously shown to act downstream of PXY, acts redundantly with WOX14 to regulate vascular cell division but that neither gene regulates vascular organisation. Moreover, they identify an interaction between PXY and the receptor kinase ERECTA that affects the organisation of the vascular tissue but not its rate of cell division. The researchers propose, therefore, that PXY and ERECTA signalling integrate cell division and vascular organisation in the Arabidopsis stem, thereby ensuring normal stem formation.
PLUS…
An intracellular partitioning-based framework for tissue cell polarity in plants and animals
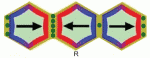 Tissue cell polarity plays a major role in plant and animal development. Coen and colleagues propose that a fundamental building block for tissue cell polarity is the process of intracellular partitioning, which can establish individual cell polarity in the absence of asymmetric cues. See the Hypothesis article on p. 2061.
Tissue cell polarity plays a major role in plant and animal development. Coen and colleagues propose that a fundamental building block for tissue cell polarity is the process of intracellular partitioning, which can establish individual cell polarity in the absence of asymmetric cues. See the Hypothesis article on p. 2061.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...
 Stem Cells Image Competition
Stem Cells Image Competition – We had our first “Journal Club on the Node” post – thanks to the University of Chicago Development, Regeneration and Stem Cell Journal Club for sharing their discussion with us. We look forward to seeing many more of these journal clubs – from U. Chicago and any other journals clubs that are interested – on the Node in the future.
– We had our first “Journal Club on the Node” post – thanks to the University of Chicago Development, Regeneration and Stem Cell Journal Club for sharing their discussion with us. We look forward to seeing many more of these journal clubs – from U. Chicago and any other journals clubs that are interested – on the Node in the future.![]() – One of Development’s sister journals, Disease Models and Mechanisms (DMM), announced the appointment of a new team of academic editors.
– One of Development’s sister journals, Disease Models and Mechanisms (DMM), announced the appointment of a new team of academic editors.![]() – Continuing with the reports from the BSDB/BSCB Joint Spring Meeting, Andrei Luchici summarised Day 1 at the meeting.
– Continuing with the reports from the BSDB/BSCB Joint Spring Meeting, Andrei Luchici summarised Day 1 at the meeting. 

 (No Ratings Yet)
(No Ratings Yet) During gametogenesis in many organisms, germ cells undergo synchronous, incomplete divisions just before meiosis that generate interconnected cell groups known as cysts, but does a cyst stage always occur during mammalian gametogenesis? Here (
During gametogenesis in many organisms, germ cells undergo synchronous, incomplete divisions just before meiosis that generate interconnected cell groups known as cysts, but does a cyst stage always occur during mammalian gametogenesis? Here ( During mitosis, ubiquitylation of the anaphase inhibitor securin by the E3 ligase anaphase-promoting complex/cyclosome (APC/C) targets securin for proteolysis, which triggers sister chromatid separation. Securin is also implicated in meiotic progression. Thus, during both mitosis and meiosis, securin expression must be tightly regulated to enable its timely action. On
During mitosis, ubiquitylation of the anaphase inhibitor securin by the E3 ligase anaphase-promoting complex/cyclosome (APC/C) targets securin for proteolysis, which triggers sister chromatid separation. Securin is also implicated in meiotic progression. Thus, during both mitosis and meiosis, securin expression must be tightly regulated to enable its timely action. On  Evolution of the capacity to form appendages (secondary outgrowths from the principal embryonic axes) potentiated the diversification of animal body plans. The basic mechanisms that underlie appendage growth in bilaterian model systems have been identified but little is known about appendage development in pre-bilaterians. Now, on
Evolution of the capacity to form appendages (secondary outgrowths from the principal embryonic axes) potentiated the diversification of animal body plans. The basic mechanisms that underlie appendage growth in bilaterian model systems have been identified but little is known about appendage development in pre-bilaterians. Now, on  The apical membrane of apical progenitors (APs) in the developing mammalian cortex is involved in cell-cell and cell-extracellular matrix (ECM) adhesion events that maintain the integrity of the developing neuroepithelium. Apical adhesion is important for several aspects of neural development but its regulation is poorly understood. Here (
The apical membrane of apical progenitors (APs) in the developing mammalian cortex is involved in cell-cell and cell-extracellular matrix (ECM) adhesion events that maintain the integrity of the developing neuroepithelium. Apical adhesion is important for several aspects of neural development but its regulation is poorly understood. Here ( The actin-bundling protein fascin 1 is expressed at specific times and places during development, and is often expressed in tumours. Fascin 1 has a clear role in cell migration in vitro but what is its role in vivo? To answer this question, Laura Machesky and co-workers have been studying the role of fascin 1 in the mouse melanocyte lineage and in human melanoma cells (
The actin-bundling protein fascin 1 is expressed at specific times and places during development, and is often expressed in tumours. Fascin 1 has a clear role in cell migration in vitro but what is its role in vivo? To answer this question, Laura Machesky and co-workers have been studying the role of fascin 1 in the mouse melanocyte lineage and in human melanoma cells ( Plant vascular tissue is derived from the cambium and procambium, which are meristems that contain long thin stem cells that divide down their long axis. These highly organised divisions produce characteristic files of cells in the xylem and ensure spatial separation of the xylem and phloem. The ligand-receptor pair CLE41-PXY regulates vascular cell division, vascular organisation and xylem differentiation. Now, on
Plant vascular tissue is derived from the cambium and procambium, which are meristems that contain long thin stem cells that divide down their long axis. These highly organised divisions produce characteristic files of cells in the xylem and ensure spatial separation of the xylem and phloem. The ligand-receptor pair CLE41-PXY regulates vascular cell division, vascular organisation and xylem differentiation. Now, on  Tissue cell polarity plays a major role in plant and animal development. Coen and colleagues propose that a fundamental building block for tissue cell polarity is the process of intracellular partitioning, which can establish individual cell polarity in the absence of asymmetric cues. See the Hypothesis article on p.
Tissue cell polarity plays a major role in plant and animal development. Coen and colleagues propose that a fundamental building block for tissue cell polarity is the process of intracellular partitioning, which can establish individual cell polarity in the absence of asymmetric cues. See the Hypothesis article on p.  (8 votes)
(8 votes)




