In this second part of my BSDB/BSCB spring meeting report, I will attempt to paint a picture of the scientific content of the conference in broad brush strokes. I have grouped talks into overarching topics and will highlight some of my favourites in each category.
Epigenetics
It is only on rare occasions that a presentation makes you go “wow” and forces you to reconsider your thinking about the most fundamental element of our bodies, the cell, and how it works. I experienced one of these moments as I was listening to Mike White (University of Manchester, UK) talk about his work on cell to cell heterogeneity. He presented fascinating live imaging data demonstrating that several molecules of the NFkB pathway periodically change location within the cell at distinct rates. Based on this, he suggested that the position of proteins could be just as important in determining the cellular response to a stimulus as the signal itself. This would add a whole new dimension to consider in the study of cell biology, which seems both exciting and daunting at the same time.
I was also particularly impressed by Martin Howard’s (John Innes Centre, UK) research, which combines experimentation and mathematical modelling to investigate epigenetics in plants. By equating histone modifications with a binary system, his group found that genes can only exist in a state of silenced (“0”) or active (“1”) state. What seems like a smooth transition from one state to the other at an organismal level is in reality caused by a change in the fraction of cells that are “0” or “1”. He also showed preliminary experimental data to support this model. Granted, the main reason I was so awed by this talk was most likely my limited understanding of mathematics, much less mathematical modelling. Regardless, I believe it illustrated the value of using mathematics to formulate elaborate hypotheses which can then be tested and validated through laboratory science.
Other recurring topics throughout the meeting included RNAi and epigenetic reprogramming in early development. Barabara Pernaute (National Institute of Medical Research, UK) presented her work on the role of miRNAs in naive embryonic stem cells compared to epiblast cells in post-implantation embryos. Similarly, Juriaan Holzenspies’ (University of Copenhagen, Denmark) presentation focused on histone marks and RNA polymerase phosphorylation states in blastocyst and epiblast. As part of the post-graduate symposium, Siyao Wang (University of Manchester, UK) presented her PhD research on the role of MLL in germ line cells of the C. elegans embryos and Richard Kaschula (University of Sussex, UK) talked about ubx-targeting miRNAs in Drosophila. Finally, Danesh Moazed (Harvard Medical School, USA) focused on his work into the mechanisms of RNAi-mediated silencing, specifically the signals initiating this process and the machinery which retains siRNA at its target site.
Stem cells, regeneration and cancer
Upon browsing the preliminary programme of speakers about two weeks before the start of the meeting, I noticed with great delight that a number of lectures would be focused on regeneration. I especially looked forward to Elly Tanaka’s (Technische Universitaet / Center for Regenerative Therapies Dresden, Germany) talk on limb regeneration in two species of salamander, axolotl and newt. She highlighted some fundamental differences in the mechanisms of limb replacement in these two organisms after amputation. I was especially struck, and perhaps a little confused, by the fact that axolotl and newt both belong to the order of salamanders, yet seem to use different mechanisms to regenerate lost appendages.
Focusing in particular on skin, Ben Simons (University of Cambridge, UK) talked about his inquiry into the mechanisms by which skin stem cells maintain their tissue. His lab’s previous work on this topic challenged the accepted theory of skin maintenance at the time. As he explained the validation and alternate experiments they carried out to confirm that their hypothesis was substantiated by the evidence, I became more and more impressed with his work. I felt that it needed a lot of confidence and courage to challenge long-standing theories and a great amount of perseverance to convince other people of the validity of your research. He also talked about the role these skin stem cells in injury repair and in the progression from papilloma to cancer.
With great relevance to my own work, I was also excited to hear about Jyotsna Dhawan’s (National Centre for Biological Sciences, India) research into the factors which mediate reversible quiescence in muscle progenitor cells. She proposed a completely new way of thinking about the G0 stage in cell cycle, suggesting that quiescent stem cells are poised for proliferation or differentiation rather than simply lying dormant. I find particularly intriguing that this may not only be the case in muscle progenitors, but also in other tissue stem cells which remain quiescent in the absence of activation stimuli. She also implicated chromatin-modulating proteins, which may act as a molecular switch.
Further talks included Cathrin Brisken (Ecole polytechnique federale de Lausanne, Switzerland), who presented hear team’s research on the role of progesterone in adult mammary gland maturation and also addressed the significance of hormone mimetics in breast cancer. Moreover, both Anna Bigas (Institut Hospital del Mar d’Investigacions Mediques, Spain) and Sarah Bray (University of Cambridge, UK) talked about the role of Notch signalling in blood cell precursor specification in mouse and Drosophila, respectively.
Patterning and early development
Another highlight for me was Olivier Pourquie’s plenary lecture about the generation and differentiation of the paraxial mesoderm. I had already been familiar with several aspects of his work, but I was surprised to hear a wealth of new findings which looked at the paraxial mesoderm from as many angles as possible. He addressed the oscillatory expression of several genes as somitogenesis takes place as well as remarkable metabolic signatures which differ between anterior and posterior paraxial mesoderm. Further, he spoke about his quest to differentiate induced pluripotent stem cells into paraxial mesoderm in vitro.
The use of unconventional organisms to get to the bottom of evolutionary questions has always fascinated and inspired me. I was thus very interested when listening to Patrick Lemaire (Centre de Recherche de Biochimie MacromoléculaireMontpellier, France) and Robb Krumlauf (Stowers Institute for Medical Research, USA). Patrick presented his work using different genera and species of ascidians, with which he investigates how morphological characteristics can stay stable across time despite major genomic changes. Robb talked about his lab’s research into the evolutionary origins of the Hox clusters using the lamprey. In both their presentations, the speakers highlighted the problems of working with such unconventional organisms and I was particularly intrigued by the fact that many of the conventional techniques in molecular biology can be adapted for these exotic organisms.
Additional talks looked at the migration of precursor cells and their differentiation. Stephen Fleenor (University of Oxford, UK) presented his PhD project on the role of G-coupled proteins in craniofacial ganglion precursor proliferation, migration and differentiation. Moreover, James McColl talked about the role of several signalling molecules in the migration of heart progenitor cells. Lastly, Ferenc Mueller (University of Birmingham, UK) addressed the complexity of promoters and enhancers, which his team investigates through the maternal-zygotic transition of transcription in the early embryo.
“Other”
In this last section, I will focus on several talks which I have not managed to assign any particular topic to. One of the last talks of the meeting was also one I considered among the most compelling. Gero Miesenbock (University of Oxford, UK) presented his work on the role of circadian and homeostatic components in sleep control in a most captivating manner. Using a Drosophila model with sleep deficits, he described his team’s experiments step by step to expose the cause of insomnia. I particularly appreciated the use of a wide range of techniques, including molecular biology, optogenetics and electrophysiology.
As part of the post-graduate symposium, Daniel Hayward (University of Exeter, UK) focused on his research on centrosome-mediated and chromatin-mediated microtubule nucleation. With a focus on moesin and actin, Nelio Rodriguez (University College London UK) presented his work on cell shape changes throughout the cell cycle. Finally, Andrei Luchici (University College London, UK) explored the role of forces in contact inhibition.
All in all…
In my opinion, the BSDB/BSCB spring meeting 2013 was a great success, with a wealth of fascinating talks and posters along with great social events which stimulated conversation and networking. I feel like I have seen and heard a lot of interesting, novel and exciting science and have had the privilege of listening to many inspiring people. This being my first “big” conference experience, I couldn’t have asked for a better meeting to go to and I am already looking forward to next year’s BSDB/BSCD spring meeting.
 (1 votes)
(1 votes)
 Loading...
Loading...
 This piece marks the first in what we hope will be an irregular series of journal club reports from the Chicago group, and we’d like to find other developmental biology or stem cell journal clubs out there interested in writing for the Node. After all the effort of reading, analysing and discussing a paper for a journal club, we’d like to give you the opportunity of sharing that discussion with a wider community, and not just with your immediate colleagues.
This piece marks the first in what we hope will be an irregular series of journal club reports from the Chicago group, and we’d like to find other developmental biology or stem cell journal clubs out there interested in writing for the Node. After all the effort of reading, analysing and discussing a paper for a journal club, we’d like to give you the opportunity of sharing that discussion with a wider community, and not just with your immediate colleagues.

 (10 votes)
(10 votes)![Figure 1 Schematic of the clock model as proposed by Thorogood (1991). (A) The bold arrow represents the timing of the AER-to-AF transition in the developmental process. (B-D) Hypothesized representations of fin/limb development in the clock model (above) with endochondral skeletal patterns of the fin/limb (below,). (B) Fin development in a teleost, demonstrating a short period of time with AER signaling prior to the AER-to-AF transition. (C) Fin development in lobe-finned fishes, showing a longer relative time with AER signaling prior to AF transformation. (D) Limb development in a tetrapod, in which AER signaling persists throughout limb development. Figure modified from Yano et al. [3]; based on Thorogood [2]; with fossil form representations in C-D from Long et al. [4].](https://thenode.biologists.com/wp-content/uploads/2013/04/node1.png)
![Figure 2 - Repeated apical fold removal caused excessive elongation of the endoskeletal region compared to control (non-removal) fin. Zebrafish larva (7 days post-fertilization) after AF removal was performed three times on the left side pectoral fin bud; right side is control fin. Black brackets indicate the endoskeletal region. Scale bar: 200µm. From Yano et al., [3].](https://thenode.biologists.com/wp-content/uploads/2013/04/node2.png)
 (4 votes)
(4 votes) (No Ratings Yet)
(No Ratings Yet) The thymus is the primary organ responsible for generating T cells. Although thymus development has been studied in mice, little is known about how the human thymus develops. Here (
The thymus is the primary organ responsible for generating T cells. Although thymus development has been studied in mice, little is known about how the human thymus develops. Here ( During embryogenesis, the anterior-posterior (AP) and dorsal-ventral (DV) axes are specified by the activity of key signalling pathways. FGF, Wnt and retinoic acid together pattern the AP axis: high activity defines more posterior tissues, which are specified later in development than anterior tissues. The BMP pathway specifies ventral fate; low BMP activity defines dorsal. Whether and how these pathways intersect to coordinate patterning of the two axes is poorly understood. On
During embryogenesis, the anterior-posterior (AP) and dorsal-ventral (DV) axes are specified by the activity of key signalling pathways. FGF, Wnt and retinoic acid together pattern the AP axis: high activity defines more posterior tissues, which are specified later in development than anterior tissues. The BMP pathway specifies ventral fate; low BMP activity defines dorsal. Whether and how these pathways intersect to coordinate patterning of the two axes is poorly understood. On 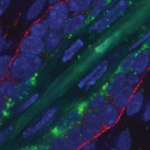 MicroRNAs are important for the regulation of gene expression in a vast array of processes. In the skin, miR-203 has been shown to be crucial for the proper differentiation of the interfollicular progenitor cells, although the specific mechanism of this has remained elusive. In this issue (
MicroRNAs are important for the regulation of gene expression in a vast array of processes. In the skin, miR-203 has been shown to be crucial for the proper differentiation of the interfollicular progenitor cells, although the specific mechanism of this has remained elusive. In this issue ( The correct establishment of adaxial-abaxial patterning is crucial for leaf expansion and growth. The AUXIN RESPONSE FACTOR (ARF) family of proteins are key determinants of organ symmetry and abaxial patterning in Arabidopsis thaliana and are subject to complex regulatory control at both the transcriptional and translational level. Here (
The correct establishment of adaxial-abaxial patterning is crucial for leaf expansion and growth. The AUXIN RESPONSE FACTOR (ARF) family of proteins are key determinants of organ symmetry and abaxial patterning in Arabidopsis thaliana and are subject to complex regulatory control at both the transcriptional and translational level. Here ( In the C. elegans embryo, anterior-posterior polarity is defined at the one-cell stage, via asymmetric and reciprocal localisation of cortex-associated PAR protein complexes: PAR-3, PAR-6 and aPKC localise to the anterior, whereas PAR-1, PAR-2 and LGL-1 are enriched at the posterior. Polarity maintenance involves mutual antagonism between the anterior and posterior complexes and may also involve CDC-42-dependent regulation of myosin activity. Kenneth Kemphues and co-workers (
In the C. elegans embryo, anterior-posterior polarity is defined at the one-cell stage, via asymmetric and reciprocal localisation of cortex-associated PAR protein complexes: PAR-3, PAR-6 and aPKC localise to the anterior, whereas PAR-1, PAR-2 and LGL-1 are enriched at the posterior. Polarity maintenance involves mutual antagonism between the anterior and posterior complexes and may also involve CDC-42-dependent regulation of myosin activity. Kenneth Kemphues and co-workers (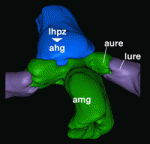 Many animal tissues maintain populations of slowly proliferating stem cells that contribute to tissue homeostasis and repair. In Drosophila, for example, stem cells reside throughout the midgut and within the hindgut and renal tubules. But how and when do these cells arise? Volker Hartenstein and colleagues now show that Drosophila gut progenitors migrate across tissue boundaries and adopt the fate of the organ in which they come to reside (
Many animal tissues maintain populations of slowly proliferating stem cells that contribute to tissue homeostasis and repair. In Drosophila, for example, stem cells reside throughout the midgut and within the hindgut and renal tubules. But how and when do these cells arise? Volker Hartenstein and colleagues now show that Drosophila gut progenitors migrate across tissue boundaries and adopt the fate of the organ in which they come to reside ( Orientation of the cell division axis is essential for both symmetric cell divisions and for the asymmetric distribution of fate determinants during, for example, stem cell divisions. Lu and Johnston review both the well-established spindle orientation pathways and recently identified regulators to provide a updated view of how positioning of the mitotic spindle occurs. See the
Orientation of the cell division axis is essential for both symmetric cell divisions and for the asymmetric distribution of fate determinants during, for example, stem cell divisions. Lu and Johnston review both the well-established spindle orientation pathways and recently identified regulators to provide a updated view of how positioning of the mitotic spindle occurs. See the 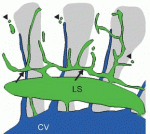 New insights into lymphatic vascular development have recently been achieved thanks to the use of alternative model systems, new molecular tools, novel imaging technologies and a growing interest in the role of lymphatic vessels in human disorders. Here. Hogan and colleagues review the most recent advances in lymphatic vascular development, with a major focus on mouse and zebrafish model systems. See the
New insights into lymphatic vascular development have recently been achieved thanks to the use of alternative model systems, new molecular tools, novel imaging technologies and a growing interest in the role of lymphatic vessels in human disorders. Here. Hogan and colleagues review the most recent advances in lymphatic vascular development, with a major focus on mouse and zebrafish model systems. See the