This book review originally appeared in Development. Tatsumi Hirata reviews “Neuronal Guidance: The Biology of Brain Wiring ” (Edited by Marc Tessier-Lavigne and Alex L. Kolodkin).
Book info:
Neuronal Guidance: The Biology of Brain Wiring. Edited by Marc Tessier-Lavigne, Alex L. Kolodkin. Cold Spring Harbor Laboratory Press (2011) 397 pages ISBN 978-0-879698-97-3 $135 (hardback)
 As a graduate student, I learned developmental neurobiology from the Principles of Neural Development (Purves and Lichtman, 1985). This masterpiece textbook taught me the fundamental concepts of how neurons are interconnected, vividly described the historical experimental basis, and introduced me to the exciting field of developmental biology. However, this book is obviously out of date today, particularly since a great number of guidance molecules and pathways have been identified in the meantime, so I have had to look for a good alternative for graduate students and postdocs interested in this field. Neuronal Guidance: The Biology of Brain Wiring completely fulfilled my expectations.
As a graduate student, I learned developmental neurobiology from the Principles of Neural Development (Purves and Lichtman, 1985). This masterpiece textbook taught me the fundamental concepts of how neurons are interconnected, vividly described the historical experimental basis, and introduced me to the exciting field of developmental biology. However, this book is obviously out of date today, particularly since a great number of guidance molecules and pathways have been identified in the meantime, so I have had to look for a good alternative for graduate students and postdocs interested in this field. Neuronal Guidance: The Biology of Brain Wiring completely fulfilled my expectations.
As this book specifically focuses on neural guidance, it might not be an appropriate textbook for undergraduate students with a broad interest in developmental biology, but it will provide a very good read for postgraduates who have some experience in neurobiology or who are keen to have a research career in this or related fields. It will also be very useful for senior investigators wanting to catch up with the latest data in the field.
Following a brief primer written by the editors, Marc Tessier-Lavigne and Alex L. Kolodkin, the book is organized into 19 chapters, each contributed by experts on the topic. Initially, I was somewhat afraid that this style might make the book a selective collection of highly specialized reviews, but, thanks to the thoughtful choice of topics and contributors, the book is, in fact, much more comprehensive than I anticipated.
The chapters are grouped into three sections, each on a different aspect of neural guidance. The first chapter, by Jonathan Raper and Carol Mason, is entitled ‘Cellular strategies of axon pathfinding’, and provides a good basic overview of the history of axon guidance research and the general concepts obtained therefrom. The following eight chapters in this first section describe comprehensive guidance strategies in traditional and new models of nervous systems. These include chapters on such as visual map development by David A. Feldheim and Dennis D. M. O’Leary, nervous system midline crossing by Barry J. Dickson and Yimin Zou, and dendrite and axon tiling by Wesley B. Grueber and Alvaro Sagasti. A little unexpected is the inclusion of the chapter entitled ‘Human genetic disorders of axon guidance’ by Elizabeth C. Engle, which effectively provides a fresh perspective on this topic.
The second section of the book includes five chapters dealing with intercellular signaling in neural guidance. This section is more detailed than the first part of the book, and is devoted to the introduction of many molecules and their signaling pathways, reflecting the rapid expansion of this field in recent years. Among the chapters here, ‘Signaling from axon guidance receptors’ is a clear overview presented by Greg J. Bashaw and Rüdiger Klein, and ‘Trafficking guidance receptors’ by Bettina Winckler and Ira Mellman describes a topic that is not always covered in axon guidance books and is informative to read.
The final section introduces other cellular processes that use common guidance signals. Several of these topics are already familiar to developmental neurobiologists, such as those described in the chapters on neural cell migration by Oscar Marín and colleagues, axon pruning by Pierre Vanderhaeghen and Hwai-Jong Cheng, and axon regeneration by Roman J. Giger and colleagues. These authors very successfully provide a good, broad perspective to the reader.
Overall, the selection of topics is well balanced, and the chapters are arranged to accurately represent the current scenario in research on neural guidance. Although a few chapters seem a little too specific for people with interests outside of that particular subdiscipline, most are written in a coherent holistic manner to provide a broad overview of our current understanding of neural guidance.
Some topics are repeatedly discussed in several sections of the book. This is the case with guidance molecules such as netrin, semaphorins, slits and ephrins, which function in various systems. Nevertheless, the overlap in the content between chapters is kept to a minimum. I prefer this approach, as it helps the reader to understand, in a step-by-step manner, how the nervous system is systematically constructed by multiple guidance signals. This makes the book more suitable to providing an understanding of the underlying biology, rather than comprising a thick directory of guidance molecules listed with location and time of action.
One feature that really impressed me is that most chapters devote a lot of attention to historical background and the description of general concepts that have led to the current state of research in the field. As mentioned above, several major principles of neural development were already established by 1985, when the classic text Principles of Neural Development was published, even though support in the form of molecular data was lacking at that time. Surprisingly, the subsequent identification of many guidance molecules and their signaling pathways has barely challenged these classic principles. Rather, new findings have confirmed established models, and, in fact, identification of most guidance molecules was based on general deductions from these principles. For example, the identification of ephrins was inspired by the historic transplantation experiments conducted on the retinotectal system by Roger Sperry and others: researchers looked for the magic chemoaffinity label that, according to theory, would form a ‘gradient’ over the tectum, and identified ephrins, which possessed exactly the predicted properties (Cheng et al., 1995; Drescher et al., 1995). Neuronal Guidance: The Biology of Brain Wiring pleasantly reminded me of such links between historic concepts and the latest data on molecular mechanisms. Realization of such connections will clarify, especially to newcomers in this field, the real significance of ongoing studies.
In summary, Neuronal Guidance: The Biology of Brain Wiring achieves a good balance between basic principles and leading cutting-edge research and will prove to be an excellent introductory text for students and postgraduates who are curious about this branch of developmental biology. The book is also refreshing for old hands: after reading it, I felt full of new ideas.
 (1 votes)
(1 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) As a graduate student, I learned developmental neurobiology from the Principles of Neural Development (
As a graduate student, I learned developmental neurobiology from the Principles of Neural Development ( (1 votes)
(1 votes) An interview with Roger Barker
An interview with Roger Barker A gene expression oscillator called the segmentation clock controls the periodic formation of somites in vertebrate embryos. In zebrafish, negative autoregulation of the transcriptional repressor genes her1 and her7 is thought to control the clock’s oscillations. Delays in this negative-feedback loop, including transcriptional delay (the time taken to make each her1 or her7 mRNA) should thus control the clock’s oscillation period. On
A gene expression oscillator called the segmentation clock controls the periodic formation of somites in vertebrate embryos. In zebrafish, negative autoregulation of the transcriptional repressor genes her1 and her7 is thought to control the clock’s oscillations. Delays in this negative-feedback loop, including transcriptional delay (the time taken to make each her1 or her7 mRNA) should thus control the clock’s oscillation period. On 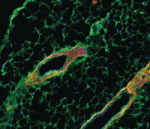 During mammary gland branching morphogenesis, mammary epithelial cells (MECs) invade the surrounding stroma. Here (
During mammary gland branching morphogenesis, mammary epithelial cells (MECs) invade the surrounding stroma. Here (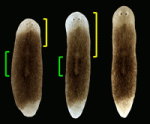 Regenerative medicine aims to replace lost or damaged tissues and organs with functional parts of the correct size and shape. To achieve this goal, we need to understand what determines the scale and form of regenerating tissues. Michael Levin and colleagues have been tackling this issue by investigating the regulation of organ size during planarian regeneration (see
Regenerative medicine aims to replace lost or damaged tissues and organs with functional parts of the correct size and shape. To achieve this goal, we need to understand what determines the scale and form of regenerating tissues. Michael Levin and colleagues have been tackling this issue by investigating the regulation of organ size during planarian regeneration (see 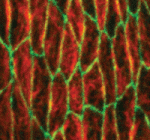 The conserved atypical cadherin Fat regulates planar cell polarity, but the mechanisms by which Fat controls cell shape and tissue organisation are unclear. Emily Marcinkevicius and Jennifer Zallen (
The conserved atypical cadherin Fat regulates planar cell polarity, but the mechanisms by which Fat controls cell shape and tissue organisation are unclear. Emily Marcinkevicius and Jennifer Zallen (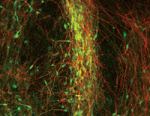 Medium-sized spiny neurons (MSNs) are the only neostriatum projection neurons and are among the first neurons to degenerate in Huntington’s disease. Here (see
Medium-sized spiny neurons (MSNs) are the only neostriatum projection neurons and are among the first neurons to degenerate in Huntington’s disease. Here (see 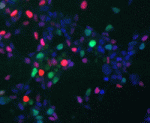 Cell-cycle progression and lineage commitment of stem cells seem to be tightly linked but it has not been possible to study this relationship in live stem cells. Now, Matthias Lutolf and colleagues (
Cell-cycle progression and lineage commitment of stem cells seem to be tightly linked but it has not been possible to study this relationship in live stem cells. Now, Matthias Lutolf and colleagues (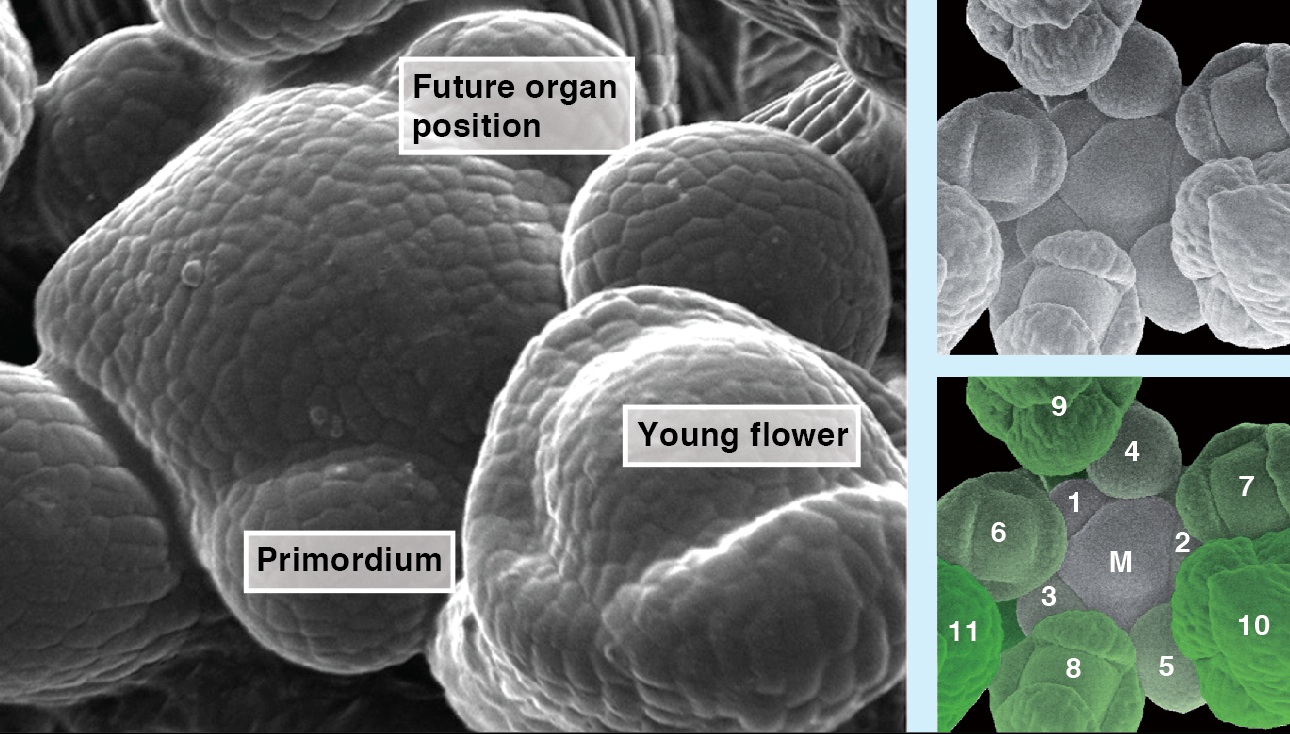 The precise arrangement of plant organs, also called phyllotaxis, has fascinated scientists from multiple disciplines. In this Issue, Jan Traas reviews new insights into the regulation of phyllotactic patterning and provides an overview of the various factors that can drive these robust growth patterns. See the Development at a Glance poster article on p.
The precise arrangement of plant organs, also called phyllotaxis, has fascinated scientists from multiple disciplines. In this Issue, Jan Traas reviews new insights into the regulation of phyllotactic patterning and provides an overview of the various factors that can drive these robust growth patterns. See the Development at a Glance poster article on p. 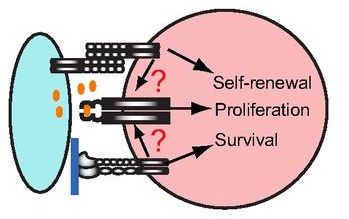 Stem cells are often anchored to their niche via adhesion molecules. However, as reviewed here by Xie and colleagues, recent studies have revealed other important roles for adhesion molecules in the regulation of stem cell function. See the Review article on p.
Stem cells are often anchored to their niche via adhesion molecules. However, as reviewed here by Xie and colleagues, recent studies have revealed other important roles for adhesion molecules in the regulation of stem cell function. See the Review article on p.