Book review: Everything you always wanted to know about auxin but were afraid to ask
Posted by Development Book Reviews, on 6 December 2012
This book review originally appeared in Development. Sabrina Sabatini reviews “Auxin Signaling: From Synthesis to Systems Biology ” (Edited by Mark Estelle, Dolf Weijers, Karin Ljung and Ottoline Leyser).
Book info:
Auxin Signaling: From Synthesis to Systems Biology. Edited by Mark Estelle, Dolf Weijers, Karin Ljung, Ottoline Leyser. Cold Spring Harbor Laboratory Press (2011) 253 pages ISBN 978-0-879698-98-0 $67.50 (hardback)
 Whether you like it or not, if you work in plant science it is of utmost importance that you know what auxin is and does, since your research will, at some point, most certainly cross the auxin path. The plant hormone auxin regulates a plethora of developmental and physiological processes in plants. At the organismal level, the differential accumulation of auxin forms gradients, which, through interactions with other hormones, regulate processes as diverse as responses to external stimuli and embryonic and postembryonic development. All of these macroscopic processes are a reflection of the changes in gene expression triggered by auxin. The book Auxin Signaling tries to provide a broad coverage of each aspect of auxin signaling, from synthesis to transport and signal transduction, along with an in-depth description of auxin’s role in various events in plant development.
Whether you like it or not, if you work in plant science it is of utmost importance that you know what auxin is and does, since your research will, at some point, most certainly cross the auxin path. The plant hormone auxin regulates a plethora of developmental and physiological processes in plants. At the organismal level, the differential accumulation of auxin forms gradients, which, through interactions with other hormones, regulate processes as diverse as responses to external stimuli and embryonic and postembryonic development. All of these macroscopic processes are a reflection of the changes in gene expression triggered by auxin. The book Auxin Signaling tries to provide a broad coverage of each aspect of auxin signaling, from synthesis to transport and signal transduction, along with an in-depth description of auxin’s role in various events in plant development.
The book opens with a chapter by Abel and Theologis on the historical context of auxin research, which provides an evocative description of the scientific journey that brought us to the present level of knowledge of auxin signaling. This section thrills the reader with the ups and downs of auxin research, and spurs them to read the rest of the book to discover where the frontline of auxin research is now. The book then divides into four sections: auxin synthesis and transport, auxin perception and cellular responses, auxin in development, and signal integration.
The second and the third sections work together to illustrate the conceptually sequential mechanisms by which auxin is synthesized, metabolized, transported and perceived by the cell, followed by the basic cellular response. The first chapter of these describes the state of our knowledge of the auxin homeostasis pathway, despite the difficulties in the quantification of auxin and its intermediates. Apart from giving an in-depth description of the pathway, the authors have included many references to the experiments behind the facts, enticing the reader to critically assess the source of information. The next two chapters provide a unique perspective on classical topics such as auxin transport and perception. In ‘Auxin transporters – why so many?’, the authors illustrate the complexity and redundancy of the auxin transport machinery in the light of evolution, offering a refreshing (and for once non-PIN-centric) point of view. The authors of the next chapter provide a very interesting structural insight into auxin perception, cleverly describing how auxin acts differently from other hormones, the application of this mechanism as a model in drug development, and, notably, a controversial idea of what truly constitutes an auxin receptor. Perrot-Rechenmann nicely concludes this section by bringing the reader from the purely molecular to the cellular level of the auxin response, as she skillfully describes all the intricate networks that regulate the two major destinies of a non-dying cell: division or differentiation.
After explaining all the main features of auxin cellular signaling, the fourth section of the book, which might interest the readers of Development most, focuses on the role of auxin in plant development. The seven chapters that constitute this section cover the classical developmental events in which auxin has been known to play a major role: embryogenesis, shoot and root meristem activity, lateral root and vascular formation, and the differentiation of reproductive organs. The added bonus of this section is an opening chapter on mathematical modeling and a closing chapter on auxin in monocotyledon (monocot) development, which nicely broaden the horizon of the reader from the classical topics of auxin-regulated plant development.
The chapter on mathematical modeling describes recent efforts made to assist research on the role of auxin in plant development. Modeling is an invaluable tool for the study of convoluted signaling pathways, such as auxin’s, in the context of complex events, such as those that occur in development. In addition to its descriptive function, mathematical modeling is, most importantly, an invaluable instrument for the generation of new hypotheses, and any researcher working in development nowadays cannot ignore it. This chapter, by explaining each model in terms that are not overly technical, gives a good basic overview of modeling in auxin biology while also representing a good starting point for those who, after reading it, want to know more.
The role of auxin in plant development is complex, but the chapters that constitute the core of this section do an excellent job at summarizing the vast literature on the topic. Moreover, each piece covers the most advanced data and theories of their topic, making this section a very good read, even, if not especially, for those that already work in the auxin field. Despite the clarity and the brief introductory remarks in all chapters, it will be better for the novice reader to have grasped the notions illustrated in the previous sections in order to get the most from these chapters.
The concluding chapter of this section not only describes the role of auxin in monocot development, but also gives a detailed overview on monocot development itself. Some plant researchers tend to focus their attention, for practical reasons, on dicots. This chapter, although explicative and comprehensive, reminds us that the comparative study of auxin function can, in the light of evolution, tell us more about auxin signaling than independent study in a single phylum.
The last section of the book deals with signal integration. The introductory chapter offers an unusual view of auxin as the currency of the plant cell. Beyond this singular metaphor, the chapter gives a broad overview of auxin pathway interaction with the light response, the circadian clock, other hormones and pathogens. The chapter that follows neatly describes all the molecular principles underlying the complexity that can be generated within the auxin pathway. In my opinion, this piece should be read before the chapters on development, as it lays fundamental concepts that would help the reader to better interpret the events covered in the previous section. This is actually easy to do, since, as can be said for the whole book, each chapter is quite autonomous and can be read independently. The remaining chapters in this section cover the interplay between the auxin pathway and the response to light, and the importance of auxin in the plant-microbe interaction. Despite the high quality of this section, I believe that it lacks chapters on the role of auxin in the response to nutrient abundance, drought and the effects of other hormones on the auxin pathway. Although hints on these topics are given throughout, dedicated chapters would have made this book truly comprehensive.
Overall, this volume suffers slightly from being a subject collection, rather than a book. In particular, even though there is a preface from the editors and an excellent opening chapter, there is no concluding chapter, which leaves the reader slightly baffled at the end of the book. This is a common problem in scientific texts, but in a volume on a topic as complex as auxin signaling, a concluding chapter would have made the work feel more like a book and less like a well-edited collection of reviews.
However, each chapter in Auxin Signaling is truly remarkable: well written and easy to follow, providing sufficiently detailed yet understandable descriptions of complex concepts, and allowing a glimpse at the experimental procedures behind them. The authors have succeeded in producing a volume that could be extremely useful for a graduate student pursuing a research career in plant biology and, most certainly, for more experienced scientists who wish to gain comprehensive knowledge of the current state of auxin research. Indeed, the book is not only descriptive, but also provides the personal perspectives of the authors and thought-provoking discussions, making it of great interest for those who work in the auxin field. Although Auxin Signaling might be too specific for an undergraduate reader, its clarity makes it a good tool for lecturers and for those curious students who could still extract important notions and concepts from it.


 (1 votes)
(1 votes) (No Ratings Yet)
(No Ratings Yet)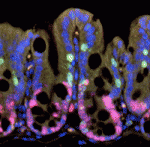 The intestinal epithelium continuously renews throughout life. Canonical Wnt signalling – a major player in this renewal – both controls intestinal epithelial proliferation and maintains intestinal stem cells. The existence of a gradient of β-catenin expression along the colonic crypt axis, with the highest β-catenin levels at the bottom where the intestinal stem cells reside, suggests that Wnt signalling might have dose-dependent roles in the colonic epithelium. On
The intestinal epithelium continuously renews throughout life. Canonical Wnt signalling – a major player in this renewal – both controls intestinal epithelial proliferation and maintains intestinal stem cells. The existence of a gradient of β-catenin expression along the colonic crypt axis, with the highest β-catenin levels at the bottom where the intestinal stem cells reside, suggests that Wnt signalling might have dose-dependent roles in the colonic epithelium. On 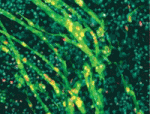 Following muscle injury, quiescent muscle stem cells (satellite cells) are activated and then proliferate and differentiate to regenerate myofibres. Wnt signalling regulates the differentiation of activated satellite cells, but what other factors control skeletal muscle regeneration? On
Following muscle injury, quiescent muscle stem cells (satellite cells) are activated and then proliferate and differentiate to regenerate myofibres. Wnt signalling regulates the differentiation of activated satellite cells, but what other factors control skeletal muscle regeneration? On  During development, cell type-specific epigenetic states must be duplicated accurately during mitosis to maintain cellular memory. The maintenance of epigenetic memory seems to involve the DNA replication machinery but is poorly understood. Here (
During development, cell type-specific epigenetic states must be duplicated accurately during mitosis to maintain cellular memory. The maintenance of epigenetic memory seems to involve the DNA replication machinery but is poorly understood. Here (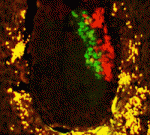 Mouse embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs) correspond to the naïve ground state of the preimplantation epiblast and the primed state of the post-implantation epiblast, respectively. The ESC state is characterised by spontaneous, reversible differences among the ESCs in their susceptibility to self-renewal and differentiation signals. Here (
Mouse embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs) correspond to the naïve ground state of the preimplantation epiblast and the primed state of the post-implantation epiblast, respectively. The ESC state is characterised by spontaneous, reversible differences among the ESCs in their susceptibility to self-renewal and differentiation signals. Here (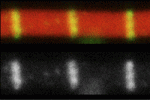 Duchenne muscular dystrophy – a lethal disease caused by dystrophin mutations – is characterised by age-dependent muscle wasting. Mario Pantoja and co-workers now report that increased intracellular sphingosine 1-phosphate (S1P) suppresses dystrophic muscle phenotypes in a Drosophila Dystrophin mutant (see
Duchenne muscular dystrophy – a lethal disease caused by dystrophin mutations – is characterised by age-dependent muscle wasting. Mario Pantoja and co-workers now report that increased intracellular sphingosine 1-phosphate (S1P) suppresses dystrophic muscle phenotypes in a Drosophila Dystrophin mutant (see  Visualising cell-cycle progression in living embryos is essential for improving our understanding of developmental processes. The fluorescent ubiquitylation-based cell-cycle indicator (Fucci), which was generated by fusing the ubiquitylation domains of Cdt1 and Geminin to different fluorescent proteins, has been used to label G1 and S/G2/M phase nuclei orange and green, respectively. Cell cycle dynamics during development have been studied in transgenic mice expressing Fucci under the control of the CAG promoter but Fucci expression levels are somewhat variable. Now, Shinichi Aizawa and colleagues (
Visualising cell-cycle progression in living embryos is essential for improving our understanding of developmental processes. The fluorescent ubiquitylation-based cell-cycle indicator (Fucci), which was generated by fusing the ubiquitylation domains of Cdt1 and Geminin to different fluorescent proteins, has been used to label G1 and S/G2/M phase nuclei orange and green, respectively. Cell cycle dynamics during development have been studied in transgenic mice expressing Fucci under the control of the CAG promoter but Fucci expression levels are somewhat variable. Now, Shinichi Aizawa and colleagues (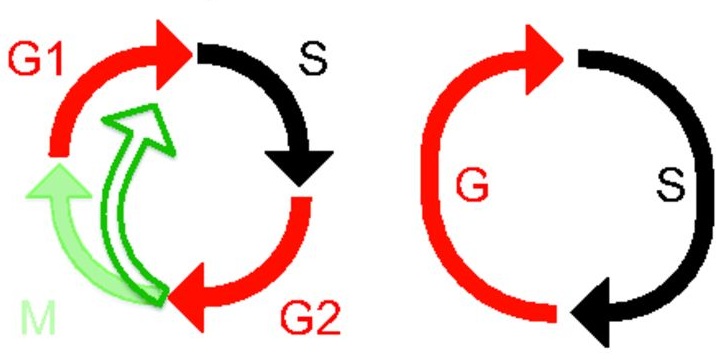 Polyploid cells have genomes that contain multiples of the typical diploid chromosome number and are found in many different organisms. As part of the “Development: The Big Picture” series, Fox and Duronio review the conserved mechanisms that control the generation of polyploidy and highlight studies that have begun to provide clues into the physiological function of polyploidy. See the Primer on p.
Polyploid cells have genomes that contain multiples of the typical diploid chromosome number and are found in many different organisms. As part of the “Development: The Big Picture” series, Fox and Duronio review the conserved mechanisms that control the generation of polyploidy and highlight studies that have begun to provide clues into the physiological function of polyploidy. See the Primer on p.  Determinants of cell polarity dictate the behaviour of many cell types during development. Recent work in yeast, worms and flies, reviewed here by Barry Thompson, has combined computer modelling with experimental analysis to reveal the mechanisms that drive the polarisation of such determinants. See the Review on p.
Determinants of cell polarity dictate the behaviour of many cell types during development. Recent work in yeast, worms and flies, reviewed here by Barry Thompson, has combined computer modelling with experimental analysis to reveal the mechanisms that drive the polarisation of such determinants. See the Review on p.  (3 votes)
(3 votes)