Here are the highlights from the current issue of Development:
Translating Apc1 loss into intestinal proliferation
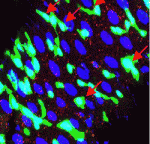 Most colorectal cancers carry inactivating mutations in adenomatous polyposis coli (APC), a negative regulator of Wnt signalling. Moreover, in mice, Apc inactivation is sufficient to drive intestinal hyperplasia. Now, Julia Cordero, Owen Sansom and colleagues use the adult Drosophila midgut to identify the signalling pathways that mediate intestinal hyperplasia following Apc loss (see p. 4524). The researchers show that the Wnt target Myc is required for Drosophila intestinal stem cell (ISC) hyperproliferation following Apc1 loss. Egfr and Jak/Stat signalling are both activated in a Myc-dependent manner following Apc1 loss, they report, and are required to drive ISC hyperproliferation. Notably, Apc1 loss in ISCs results in non-cell-autonomous upregulation of the interleukin-like ligands Upd2/3 in enterocytes and subsequent activation of Jak/Stat signalling in ISCs. These and other findings indicate that non-cell-autonomous crosstalk between the Wnt/Myc, Egfr and Jak/Stat signalling pathways regulates intestinal hyperproliferation in the Drosophila midgut following Apc1 loss and suggest that these pathways may also be activated in human colorectal cancers
Most colorectal cancers carry inactivating mutations in adenomatous polyposis coli (APC), a negative regulator of Wnt signalling. Moreover, in mice, Apc inactivation is sufficient to drive intestinal hyperplasia. Now, Julia Cordero, Owen Sansom and colleagues use the adult Drosophila midgut to identify the signalling pathways that mediate intestinal hyperplasia following Apc loss (see p. 4524). The researchers show that the Wnt target Myc is required for Drosophila intestinal stem cell (ISC) hyperproliferation following Apc1 loss. Egfr and Jak/Stat signalling are both activated in a Myc-dependent manner following Apc1 loss, they report, and are required to drive ISC hyperproliferation. Notably, Apc1 loss in ISCs results in non-cell-autonomous upregulation of the interleukin-like ligands Upd2/3 in enterocytes and subsequent activation of Jak/Stat signalling in ISCs. These and other findings indicate that non-cell-autonomous crosstalk between the Wnt/Myc, Egfr and Jak/Stat signalling pathways regulates intestinal hyperproliferation in the Drosophila midgut following Apc1 loss and suggest that these pathways may also be activated in human colorectal cancers
Maelstrom regulation of oocyte determination
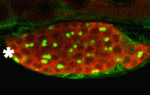 The conserved protein Maelstrom has multiple functions during Drosophila oogenesis. It is a component of the perinuclear nuage, a germline structure in which the piRNAs that are required for transposon silencing are produced. In addition, Maelstrom localised within the nucleus promotes germline stem cell differentiation by repressing expression of the microRNA miR-7. Here (p. 4505), Toshie Kai and co-workers describe a third way in which Maelstrom regulates oogenesis. They show that Maelstrom is phosphorylated in Drosophila ovaries and identify its conserved serine 138 residue as the phosphorylation site. Maelstrom phosphorylation, they report, is required for oocyte determination but not for transposon silencing or for germline stem cell differentiation. They also show that phosphorylation of Maelstrom represses the pachytene checkpoint (one of the two meiotic checkpoints in Drosophila), in part through downregulation of the checkpoint protein Sir2, and identify Polo as a kinase that mediates Maelstrom phosphorylation. Thus, the researchers propose, Polo-mediated phosphorylation of Maelstrom regulates oocyte determination in Drosophila ovaries by inactivating the pachytene checkpoint.
The conserved protein Maelstrom has multiple functions during Drosophila oogenesis. It is a component of the perinuclear nuage, a germline structure in which the piRNAs that are required for transposon silencing are produced. In addition, Maelstrom localised within the nucleus promotes germline stem cell differentiation by repressing expression of the microRNA miR-7. Here (p. 4505), Toshie Kai and co-workers describe a third way in which Maelstrom regulates oogenesis. They show that Maelstrom is phosphorylated in Drosophila ovaries and identify its conserved serine 138 residue as the phosphorylation site. Maelstrom phosphorylation, they report, is required for oocyte determination but not for transposon silencing or for germline stem cell differentiation. They also show that phosphorylation of Maelstrom represses the pachytene checkpoint (one of the two meiotic checkpoints in Drosophila), in part through downregulation of the checkpoint protein Sir2, and identify Polo as a kinase that mediates Maelstrom phosphorylation. Thus, the researchers propose, Polo-mediated phosphorylation of Maelstrom regulates oocyte determination in Drosophila ovaries by inactivating the pachytene checkpoint.
Key role for LIN28 in nucleologenesis
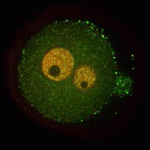 A functional nucleolus (the organelle that makes ribosome subunits) is essential for embryonic development. Nucleologenesis, which involves transformation of an inactive nucleolar precursor body (NBP) into a mature nucleolus, coincides with embryonic genome activation, which occurs at the late 2-cell stage in mouse embryos. Here (p. 4514), Edgar Vogt, Rüdiger Behr and colleagues investigate the involvement of the pluripotency factor LIN28 in nucleologenesis in mouse embryos. LIN28, which is abundantly expressed in early embryonic tissue, is an RNA-binding protein that can contribute to the reprogramming of somatic cells to pluripotent stem cells. The researchers report that LIN28 accumulates at the periphery of NPBs and mature nucleoli in preimplantation embryos and that initiation of LIN28 expression coincides with embryonic genome activation. Following LIN28 knockdown, they report, most embryos arrest between the 2- and 4-cell stages. Notably, presumptive NPB sites are not enriched with the nucleolar marker nucleophosmin in these arrested embryos. Thus, the researchers conclude, LIN28 is essential for NPB assembly and nucleologenesis during early embryonic development.
A functional nucleolus (the organelle that makes ribosome subunits) is essential for embryonic development. Nucleologenesis, which involves transformation of an inactive nucleolar precursor body (NBP) into a mature nucleolus, coincides with embryonic genome activation, which occurs at the late 2-cell stage in mouse embryos. Here (p. 4514), Edgar Vogt, Rüdiger Behr and colleagues investigate the involvement of the pluripotency factor LIN28 in nucleologenesis in mouse embryos. LIN28, which is abundantly expressed in early embryonic tissue, is an RNA-binding protein that can contribute to the reprogramming of somatic cells to pluripotent stem cells. The researchers report that LIN28 accumulates at the periphery of NPBs and mature nucleoli in preimplantation embryos and that initiation of LIN28 expression coincides with embryonic genome activation. Following LIN28 knockdown, they report, most embryos arrest between the 2- and 4-cell stages. Notably, presumptive NPB sites are not enriched with the nucleolar marker nucleophosmin in these arrested embryos. Thus, the researchers conclude, LIN28 is essential for NPB assembly and nucleologenesis during early embryonic development.
Touch-sensitive morphogenesis
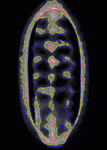 Contact inhibition of locomotion was first described more than 50 years ago by Michael Abercrombie, who reported that migrating fibroblasts change direction and migrate away from each other when they collide. Although little is known about contact inhibition in vivo, it was recently reported that contact inhibition helps to drive the uniform embryonic dispersal of Drosophila macrophages (haemocytes). Now, Graham Dunn, Brian Stramer and colleagues (p. 4555) mathematically analyse and simulate the contact repulsion dynamics of haemocytes moving along the ventral surface of the Drosophila embryo to investigate the involvement of contact inhibition in the patterning of haemocyte movements. Their simulation, which they test experimentally, reveals that the final pattern of haemocyte distribution, as well as the timing of pattern formation, can be explained by contact inhibition dynamics within the geometry of the Drosophila embryo, irrespective of external cues. This finding has broad implications for morphogenesis because it suggests that contact inhibition of locomotion can be a significant driving force for embryonic pattern formation.
Contact inhibition of locomotion was first described more than 50 years ago by Michael Abercrombie, who reported that migrating fibroblasts change direction and migrate away from each other when they collide. Although little is known about contact inhibition in vivo, it was recently reported that contact inhibition helps to drive the uniform embryonic dispersal of Drosophila macrophages (haemocytes). Now, Graham Dunn, Brian Stramer and colleagues (p. 4555) mathematically analyse and simulate the contact repulsion dynamics of haemocytes moving along the ventral surface of the Drosophila embryo to investigate the involvement of contact inhibition in the patterning of haemocyte movements. Their simulation, which they test experimentally, reveals that the final pattern of haemocyte distribution, as well as the timing of pattern formation, can be explained by contact inhibition dynamics within the geometry of the Drosophila embryo, irrespective of external cues. This finding has broad implications for morphogenesis because it suggests that contact inhibition of locomotion can be a significant driving force for embryonic pattern formation.
PBAP sets the (chromatin) boundaries
 The establishment of chromatin boundaries (transcriptional regulatory elements that counteract the spreading of silent chromatin) is essential for proper development. At these boundaries, the histone variant H3.3 replaces H3.1 in a process that, in Drosophila, involves the sequence-specific DNA-binding protein GAGA factor, the chromatin remodelling complex FACT, and the H3.3-specific chaperone HIRA. The H3.3 replacement is also likely to require an ATP-dependent remodelling factor and, on p. 4582, Susumu Hirose and colleagues identify this factor. The researchers show that GAGA factor associates with the Polybromo-associated Brm (PBAP) remodelling complex, which consists of several Trithorax group proteins, and recruits it to chromatin boundaries in Drosophila. They further show that mutations in GAGA factor, Brm and Polybromo/Bap180 disrupt H3.3 replacement and boundary function in a synergistic manner. Moreover, PBAP is needed to generate a DNase-hypersensitive site at the d1 chromatin boundary and HIRA reverses this alteration. Based on these results, the researchers propose a model for H3.3 replacement at chromatin boundaries.
The establishment of chromatin boundaries (transcriptional regulatory elements that counteract the spreading of silent chromatin) is essential for proper development. At these boundaries, the histone variant H3.3 replaces H3.1 in a process that, in Drosophila, involves the sequence-specific DNA-binding protein GAGA factor, the chromatin remodelling complex FACT, and the H3.3-specific chaperone HIRA. The H3.3 replacement is also likely to require an ATP-dependent remodelling factor and, on p. 4582, Susumu Hirose and colleagues identify this factor. The researchers show that GAGA factor associates with the Polybromo-associated Brm (PBAP) remodelling complex, which consists of several Trithorax group proteins, and recruits it to chromatin boundaries in Drosophila. They further show that mutations in GAGA factor, Brm and Polybromo/Bap180 disrupt H3.3 replacement and boundary function in a synergistic manner. Moreover, PBAP is needed to generate a DNase-hypersensitive site at the d1 chromatin boundary and HIRA reverses this alteration. Based on these results, the researchers propose a model for H3.3 replacement at chromatin boundaries.
Notch muscles in on myogenesis
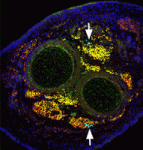 The capacity of stem cells to generate successive classes of committed progeny during development has been well-studied in some systems but is largely undefined for many tissues. Now, on p. 4536, Shahragim Tajbakhsh and co-workers investigate the role of Notch in the temporal specification potential of mouse skeletal muscle stem cells. The researchers show that Notch is active throughout development in the muscle founder stem cell population, and that expression of activated Notch (NICD) is sufficient to autonomously maintain self-renewing muscle stem/progenitor cells throughout embryogenesis, despite the absence of committed progeny, which were previously shown to be required for muscle stem cell maintenance. NICD-expressing replicating embryonic stem/progenitors respond to foetal environmental cues as development proceeds. Furthermore, the researchers report, siRNA-mediated silencing of NICD in this population promotes the temporally appropriate foetal myogenic fate, despite the expression of markers for multiple cell types. Given these results, the researchers propose that Notch signalling both maintains muscle stem cells and allows them to be receptive to specification cues throughout development.
The capacity of stem cells to generate successive classes of committed progeny during development has been well-studied in some systems but is largely undefined for many tissues. Now, on p. 4536, Shahragim Tajbakhsh and co-workers investigate the role of Notch in the temporal specification potential of mouse skeletal muscle stem cells. The researchers show that Notch is active throughout development in the muscle founder stem cell population, and that expression of activated Notch (NICD) is sufficient to autonomously maintain self-renewing muscle stem/progenitor cells throughout embryogenesis, despite the absence of committed progeny, which were previously shown to be required for muscle stem cell maintenance. NICD-expressing replicating embryonic stem/progenitors respond to foetal environmental cues as development proceeds. Furthermore, the researchers report, siRNA-mediated silencing of NICD in this population promotes the temporally appropriate foetal myogenic fate, despite the expression of markers for multiple cell types. Given these results, the researchers propose that Notch signalling both maintains muscle stem cells and allows them to be receptive to specification cues throughout development.
Plus…
The effort to make mosaic analysis a household tool
 In another ‘Development Classic’ article, Xu and Rubin describe how they set out to improve the methods used to make genetic mosaics in Drosophila and how these efforts led to their 1993 Development paper.
In another ‘Development Classic’ article, Xu and Rubin describe how they set out to improve the methods used to make genetic mosaics in Drosophila and how these efforts led to their 1993 Development paper.
See the Spotlight article on p. 4501
Book Reviews
 Our final print issue of the year includes our annual Book Review section, which highlights a number of recently published books covering topics ranging from auxin signaling, the extracellular matrix and neuronal guidance, to genomics and, more broadly, the principles that shape life. We’ll post each these Book Reviews on the Node over the coming months, but you can also view them (and reviews from previous years) here.
Our final print issue of the year includes our annual Book Review section, which highlights a number of recently published books covering topics ranging from auxin signaling, the extracellular matrix and neuronal guidance, to genomics and, more broadly, the principles that shape life. We’ll post each these Book Reviews on the Node over the coming months, but you can also view them (and reviews from previous years) here.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (3 votes)
(3 votes)

 (No Ratings Yet)
(No Ratings Yet) The past 20 years has seen a revolution in genomics. From the completion of the human genome in 2003, which took 13 years, we are well on our way to achieving the next benchmark goal of having 1000 human genomes sequenced (The 1000 Genomes Project). This endeavour will provide a deep catalogue of human genetic variation. In addition, and as part of The Genome 10K Project (which aims to sequence the genomes of 10,000 vertebrate species), 2012 saw the initial assembly of the medium ground finch (Geospiza fortis) genome, one of the iconic Galapagos finches described by Charles Darwin. The results of these genome sequencing projects are available through freely accessible public databases, thus accelerating discoveries in diverse fields of biology. With the advent of next-generation sequencing platforms, the time and cost of sequencing have dropped dramatically, making the ability to sequence the human genome in a day for less than $1000 no longer science fiction but rather an event that will happen in the immediate future. The effects of this genomics revolution are widespread, and no field of biology or medicine remains untouched by the changes in sequencing throughput. Furthermore, genomic studies are so commonly highlighted by the media that a working understanding of genomics is increasingly important in undergraduate biology education.
The past 20 years has seen a revolution in genomics. From the completion of the human genome in 2003, which took 13 years, we are well on our way to achieving the next benchmark goal of having 1000 human genomes sequenced (The 1000 Genomes Project). This endeavour will provide a deep catalogue of human genetic variation. In addition, and as part of The Genome 10K Project (which aims to sequence the genomes of 10,000 vertebrate species), 2012 saw the initial assembly of the medium ground finch (Geospiza fortis) genome, one of the iconic Galapagos finches described by Charles Darwin. The results of these genome sequencing projects are available through freely accessible public databases, thus accelerating discoveries in diverse fields of biology. With the advent of next-generation sequencing platforms, the time and cost of sequencing have dropped dramatically, making the ability to sequence the human genome in a day for less than $1000 no longer science fiction but rather an event that will happen in the immediate future. The effects of this genomics revolution are widespread, and no field of biology or medicine remains untouched by the changes in sequencing throughput. Furthermore, genomic studies are so commonly highlighted by the media that a working understanding of genomics is increasingly important in undergraduate biology education. (1 votes)
(1 votes)
 Most colorectal cancers carry inactivating mutations in adenomatous polyposis coli (APC), a negative regulator of Wnt signalling. Moreover, in mice, Apc inactivation is sufficient to drive intestinal hyperplasia. Now, Julia Cordero, Owen Sansom and colleagues use the adult Drosophila midgut to identify the signalling pathways that mediate intestinal hyperplasia following Apc loss (see
Most colorectal cancers carry inactivating mutations in adenomatous polyposis coli (APC), a negative regulator of Wnt signalling. Moreover, in mice, Apc inactivation is sufficient to drive intestinal hyperplasia. Now, Julia Cordero, Owen Sansom and colleagues use the adult Drosophila midgut to identify the signalling pathways that mediate intestinal hyperplasia following Apc loss (see  The conserved protein Maelstrom has multiple functions during Drosophila oogenesis. It is a component of the perinuclear nuage, a germline structure in which the piRNAs that are required for transposon silencing are produced. In addition, Maelstrom localised within the nucleus promotes germline stem cell differentiation by repressing expression of the microRNA miR-7. Here (
The conserved protein Maelstrom has multiple functions during Drosophila oogenesis. It is a component of the perinuclear nuage, a germline structure in which the piRNAs that are required for transposon silencing are produced. In addition, Maelstrom localised within the nucleus promotes germline stem cell differentiation by repressing expression of the microRNA miR-7. Here ( A functional nucleolus (the organelle that makes ribosome subunits) is essential for embryonic development. Nucleologenesis, which involves transformation of an inactive nucleolar precursor body (NBP) into a mature nucleolus, coincides with embryonic genome activation, which occurs at the late 2-cell stage in mouse embryos. Here (
A functional nucleolus (the organelle that makes ribosome subunits) is essential for embryonic development. Nucleologenesis, which involves transformation of an inactive nucleolar precursor body (NBP) into a mature nucleolus, coincides with embryonic genome activation, which occurs at the late 2-cell stage in mouse embryos. Here ( Contact inhibition of locomotion was first described more than 50 years ago by Michael Abercrombie, who reported that migrating fibroblasts change direction and migrate away from each other when they collide. Although little is known about contact inhibition in vivo, it was recently reported that contact inhibition helps to drive the uniform embryonic dispersal of Drosophila macrophages (haemocytes). Now, Graham Dunn, Brian Stramer and colleagues (
Contact inhibition of locomotion was first described more than 50 years ago by Michael Abercrombie, who reported that migrating fibroblasts change direction and migrate away from each other when they collide. Although little is known about contact inhibition in vivo, it was recently reported that contact inhibition helps to drive the uniform embryonic dispersal of Drosophila macrophages (haemocytes). Now, Graham Dunn, Brian Stramer and colleagues ( The establishment of chromatin boundaries (transcriptional regulatory elements that counteract the spreading of silent chromatin) is essential for proper development. At these boundaries, the histone variant H3.3 replaces H3.1 in a process that, in Drosophila, involves the sequence-specific DNA-binding protein GAGA factor, the chromatin remodelling complex FACT, and the H3.3-specific chaperone HIRA. The H3.3 replacement is also likely to require an ATP-dependent remodelling factor and, on
The establishment of chromatin boundaries (transcriptional regulatory elements that counteract the spreading of silent chromatin) is essential for proper development. At these boundaries, the histone variant H3.3 replaces H3.1 in a process that, in Drosophila, involves the sequence-specific DNA-binding protein GAGA factor, the chromatin remodelling complex FACT, and the H3.3-specific chaperone HIRA. The H3.3 replacement is also likely to require an ATP-dependent remodelling factor and, on  The capacity of stem cells to generate successive classes of committed progeny during development has been well-studied in some systems but is largely undefined for many tissues. Now, on
The capacity of stem cells to generate successive classes of committed progeny during development has been well-studied in some systems but is largely undefined for many tissues. Now, on  In another ‘Development Classic’ article, Xu and Rubin describe how they set out to improve the methods used to make genetic mosaics in Drosophila and how these efforts led to their 1993 Development paper.
In another ‘Development Classic’ article, Xu and Rubin describe how they set out to improve the methods used to make genetic mosaics in Drosophila and how these efforts led to their 1993 Development paper. Our final print issue of the year includes our annual Book Review section, which highlights a number of recently published books covering topics ranging from auxin signaling, the extracellular matrix and neuronal guidance, to genomics and, more broadly, the principles that shape life. We’ll post each these Book Reviews on the Node over the coming months, but you can also view them (and reviews from previous years)
Our final print issue of the year includes our annual Book Review section, which highlights a number of recently published books covering topics ranging from auxin signaling, the extracellular matrix and neuronal guidance, to genomics and, more broadly, the principles that shape life. We’ll post each these Book Reviews on the Node over the coming months, but you can also view them (and reviews from previous years)