BSDB Gurdon Studentship Report – Raphael Schettler
Posted by Raphael Schettler, on 12 December 2022
As a BSc Biomedicine Student at the University of East Anglia, I have had the pleasure of working in the Grocott Lab for eight weeks during my summer holidays.
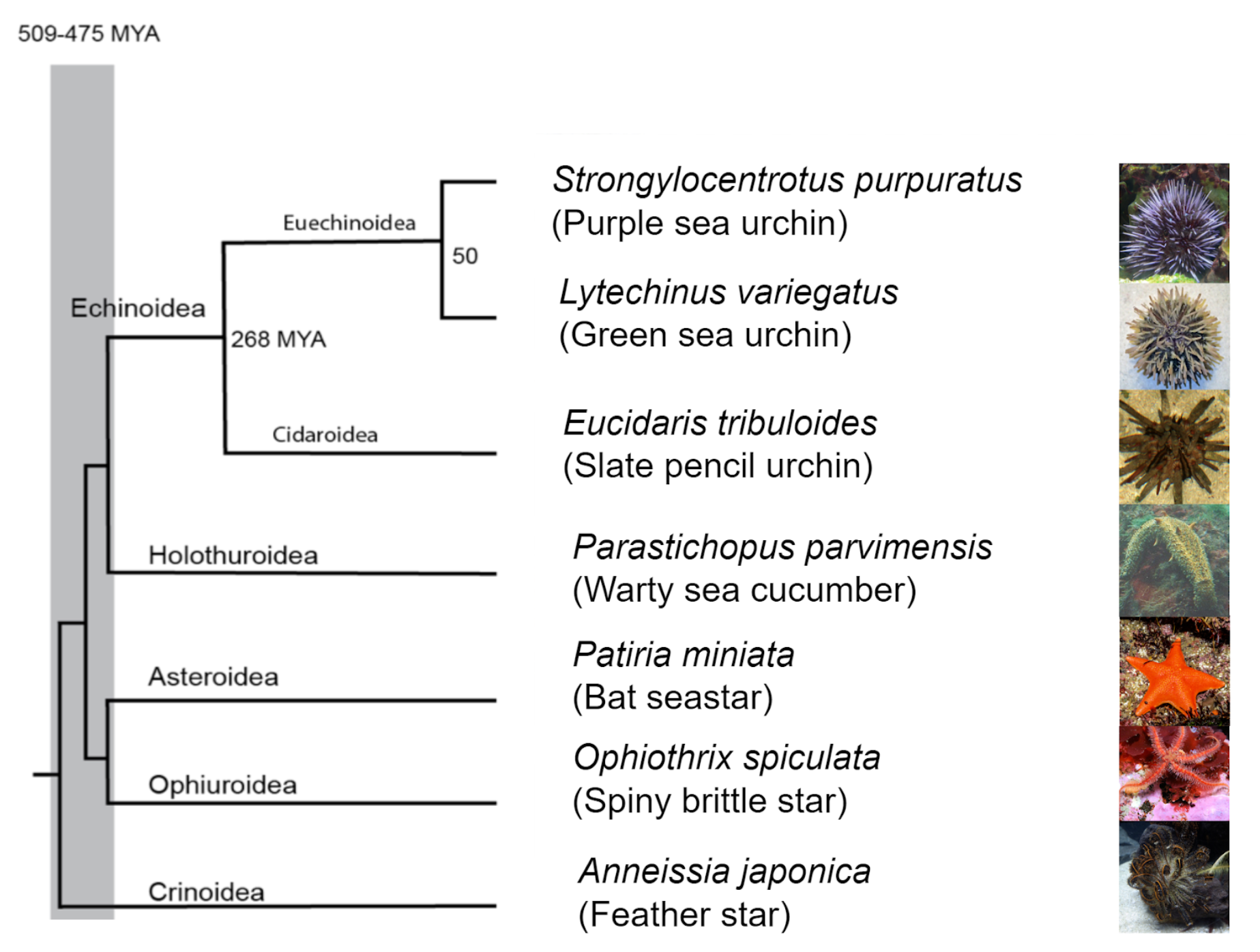
The Grocott Lab is part of the Norwich Developmental Biology and Stem Cell Network. It is researching the development of the eye, utilising chickens as a model organism. The team envisions the lab to work in three main areas: model organism work, computer modelling and human stem cell models. Figure 1 below shows the current Grocott Lab Team.

The title of my project was “Using Myc genes to search for optic vesicle progenitors”. Congenital malformations are a leading cause of infant death. Holoprosencephaly is a spectrum of related conditions in which the two sides of the brain are fused together. In extreme cases this results in a condition termed Cyclopia, where only one eye forms. The co-expression of c-Myc and N-Myc within the anterior neural folds may define paired growth fields that contribute to the outgrowth of optic vesicles. We hypothesised that the knockdown of one or both Myc genes in the anterior neural folds may result in impaired optic vesicle outgrowth and thus holoprosencephaly. This was inspired by reports that Myc genes regulate the growth of the neural retina from the optic cup lip at later stages of eye development.
Using validated antisense morpholine oligonucleotides, I knocked down the N-Myc and C-Myc genes in developing chicken embryos to assess their impact on optic vesicle outgrowth and develop an understanding of the function of both of these genes.
A typical day in the lab would include isolating embryos from eggs, preparing different chemical solutions needed to hydrate the chicken embryo, electroporation of the embryos with the morpholino (nucleic acid chains with a synthetic backbone, not recognised by cellular enzymes) mixed with DNA and incubating embryos overnight. I gathered a variety of skills during my eight weeks with the Grocott Lab. Under the close eye of Felicitas Ramirez (a postdoc in the lab and a wonderful mentor) I learnt techniques and skills necessary to culture a chicken embryo and master the art of electroporation. Electroporation is the process of introducing charged molecules, such as DNA or FITC labelled morpholinos into cells using a pulse of electricity to open pores in the cell membranes briefly.
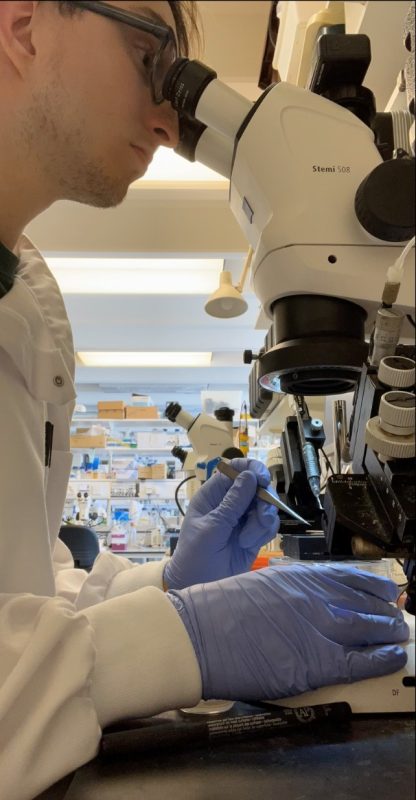
The first hurdle was isolating the embryos from eggs and getting the correct stages of development. We found that many factors such as the humidity and temperature had an effect on the stage of growth we were isolating. Figure 2 shows me at my typical workspace, the microscope, electroporating the embryos that we had isolated in the morning.
Initially, I electroporated Hamilton & Hamburger stage 3-4 embryos and cultured these for approximately 12 hours. I expected embryos to grow to stage 10, in which the optic vesicles had formed, and I could observe a phenotype. After many rounds of electroporating and with a result showing no obvious phenotype we decided to change the technique and electroporate in ovo. We kept getting a negative result and saw nothing exciting happen to the chicken embryos. Several different methods of electroporation were attempted. The only technique we found to give promising results was electroporating older embryos (stage 7-8) using the traditional method.
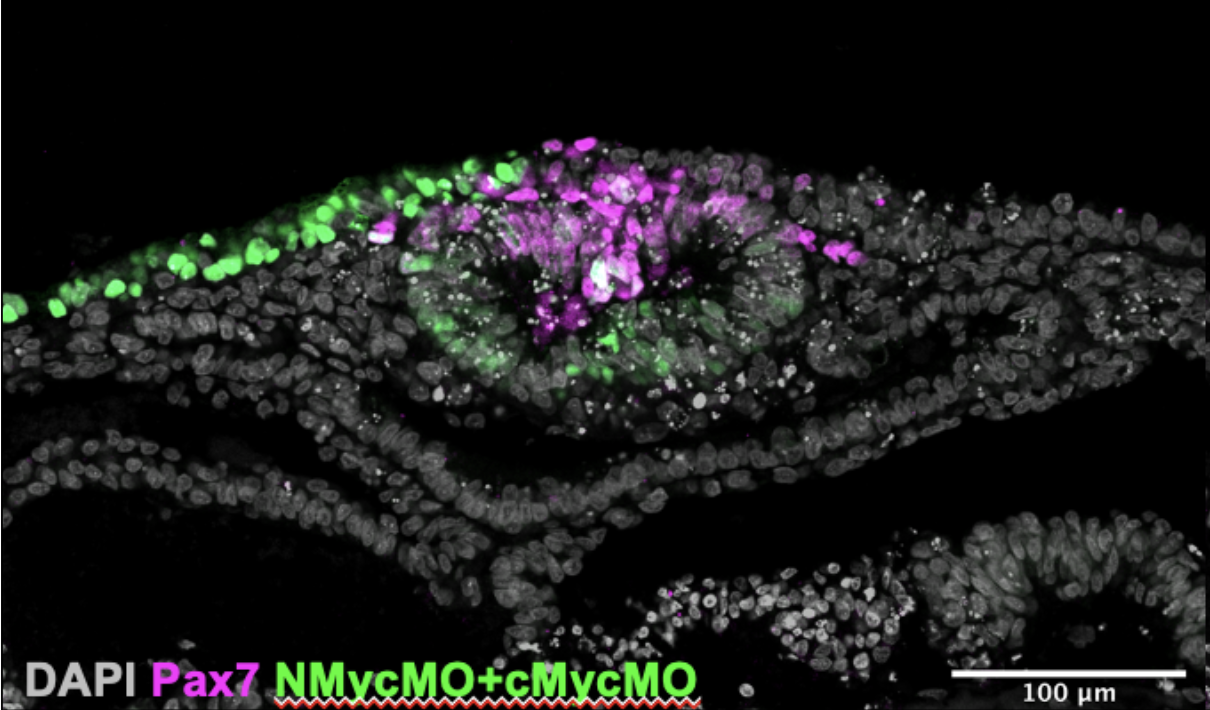
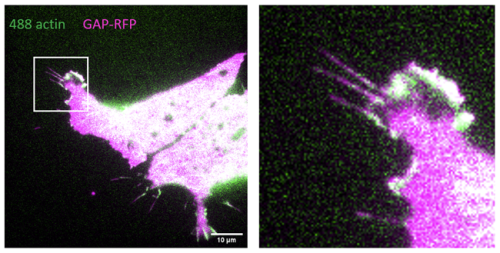
Once we had cultured the embryos and analysed them we further processed them for immuno- staining (another technique I learnt). This allowed us to look at the expression of four genes simultaneously and produced images like the one in Figure 3. We gained data that were interesting: The expression of Pax7 (a neural plate border marker) and the morpholino fluorescence was mutually exclusive. This confirms that Myc genes are important for the development of neural crest cell progenitors from the neural plate border.
From the results of our experiments we formed a new hypothesis: “The lack of obvious phenotype is because the electroporation is always mosaic and there are sufficient un-electroporated cells to compensate”. I am excited that I contributed to the project in a helpful way. Felicitas Ramirez is now continuing the project, examining the new hypothesis, and producing results which could lead to a Grocott Lab publication.
All of this would not have been possible without the support of the BSDB. I am very grateful for and appreciate the opportunity given to me. Additionally, I would like to thank Tim Grocott and his lab group for taking me in and allowing me to lend a helping hand in their research.


 (4 votes)
(4 votes)
 (No Ratings Yet)
(No Ratings Yet)