Welcome to our monthly trawl for developmental biology (and related) preprints.
The preprints this month are hosted on bioRxiv , arXiv and preprints.org – use these links to get to the section you want.
Developmental biology
Cell Biology
Modelling
Reviews
Tools & Resources
Research practice & education
Developmental biology
| Patterning & signalling
Fly embryo nuclei from Sanjuan and Bray
Membrane architecture and adherens junctions contribute to strong Notch pathway activation
Cell-autonomous generation of the wave pattern within the vertebrate segmentation clock
A protein-trap allele reveals roles for Drosophila ATF4 in photoreceptor degeneration, oocyte maturation and wing development
Interplay between cell proliferation and recruitment controls the duration of growth and final size of the Drosophila wing
Loss of imprinting of the Igf2-H19 ICR1 enhances placental endocrine capacity via sex-specific alterations in signalling pathways in the mouse
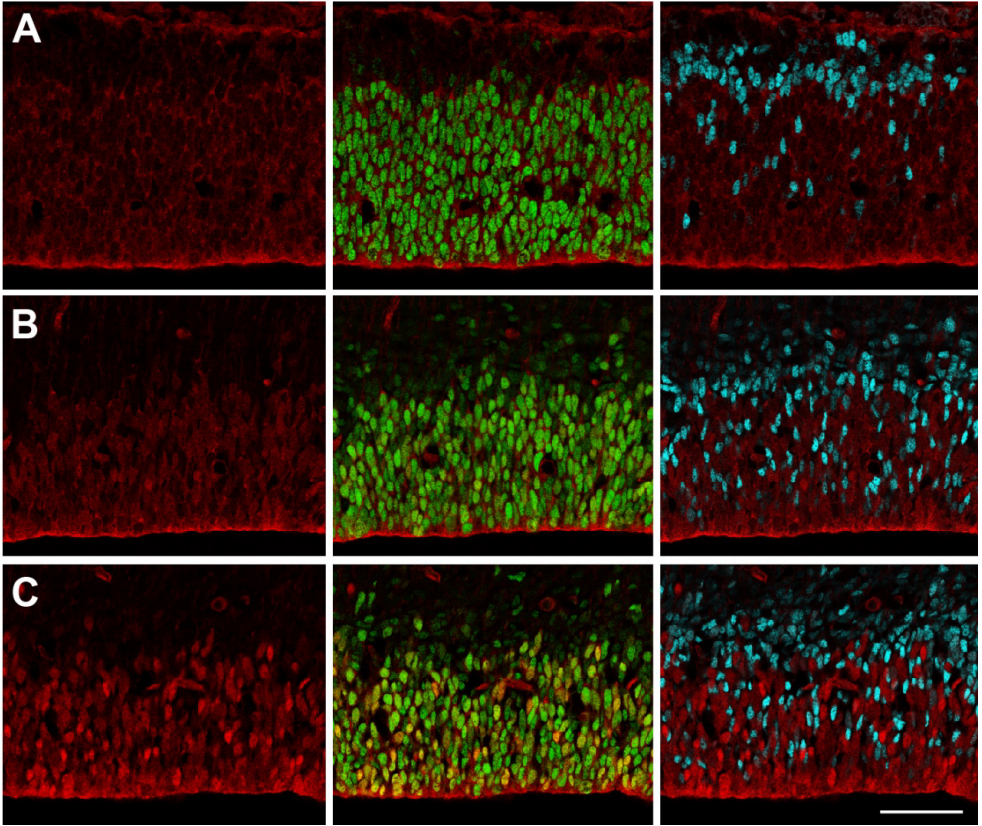
The neurogenic fate of the hindbrain boundaries: a Notch-dependent behavioral switch triggers asymmetric division of boundary stem cells
A lipid-mTORC1 nutrient sensing pathway regulates animal development by peroxisome-derived hormones
TALPID3/KIAA0586 regulates multiple aspects of neuromuscular patterning during gastrointestinal development in animal models and human
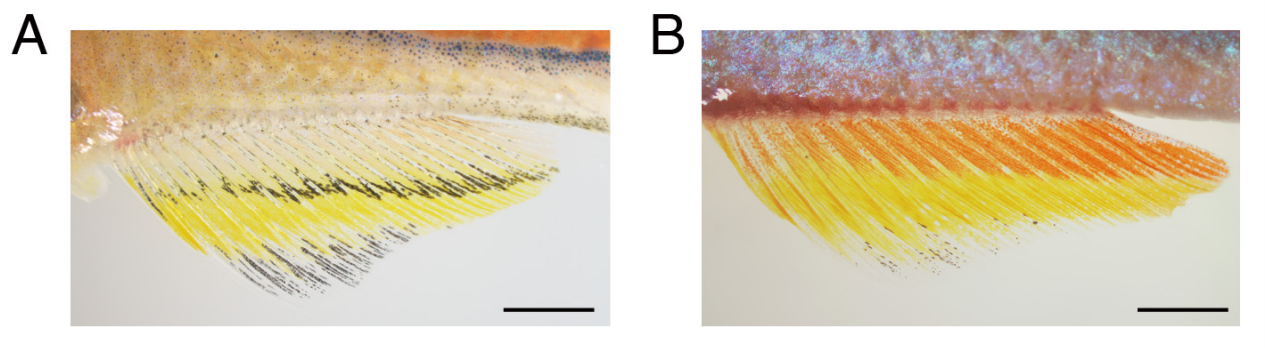
Fish fins from Huang, et al.
Development and genetics of red coloration in the zebrafish relative Danio albolineatus
A single heterozygous mutation in COG4 disrupts zebrafish early development via Wnt signaling
Buffered EGFR signaling regulated by spitz to argos expression ratio is critical for patterning the Drosophila eye
Identification of bipotent progenitors that give rise to myogenic and connective tissues in mouse
Evidence of wiring development processes from the connectome of adult Drosophila
Kap-β2/Transportin mediates β-catenin nuclear transport in Wnt signaling
The sperm protein SPACA4 is required for efficient fertilization in mice
Lyl-1 regulates primitive macrophages and microglia development
Generation and timing of graded responses to morphogen gradients
EOMES is responsible for WNT memory and can substitute for WNT in mesendoderm specification
Fly salivary glands from Du, et al.
GPI-anchored FGF directs cytoneme-mediated bidirectional signaling to self-regulate tissue-specific dispersion
Oligodendrocyte precursor cells prune axons in the mouse neocortex
Network instability dynamics drive a transient bursting period in the developing hippocampus in vivo
Callosal projections in Martín-Fernández, et al.
Role of Nrp1 in controlling cortical interhemispheric circuits
Co-option of local and systemic immune responses by the hormonal signalling system triggering metamorphosis
Mouse cortices from Lavado, et al.
YAP/TAZ Maintain the Proliferative Capacity and Structural Organization of Radial Glial Cells During Brain Development
Differential regulation of developmental stages supports a linear model for C. elegans postembryonic development
Ribosome protein mutant cells rely on the GR64 cluster of gustatory receptors for survival and proteostasis in Drosophila
Symmetry breaking in the female germline cyst
Rbfox1 is required for myofibril development and maintaining fiber-type specific isoform expression in Drosophila muscles
Autocrine regulation of adult neurogenesis by the endocannabinoid 2-arachidonoylglycerol (2-AG)
Size-dependent protein segregation creates a spatial switch for Notch signaling and function
A natural transdifferentiation event involving mitosis is empowered by integrating signaling inputs with conserved plasticity factors
Osteoblast cell death triggers a pro-osteogenic inflammatory response regulated by reactive oxygen species and glucocorticoid signaling in zebrafish
Mesothelial cells are not a source of adipocytes in mice
Key Promoter Region of Wnt4 response to FSH and Genetic Effect on Several Production Traits of Its Mutations in Chicken
Mouse heads from Shull, et al.
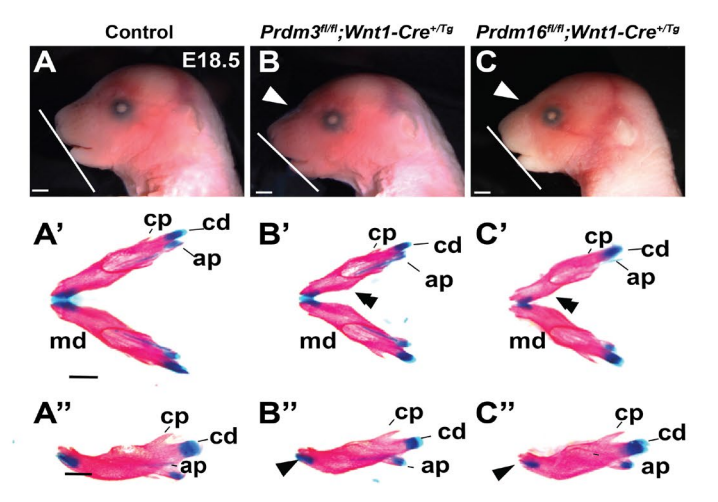
PRDM proteins control Wnt/β-catenin activity to regulate craniofacial chondrocyte differentiation
Embryonic hyperglycemia perturbs the development of specific retinal cell types, including photoreceptors
Effects of gestational age at birth on perinatal structural brain development in healthy term-born babies
Frog axonal projections in Weiss, et al.
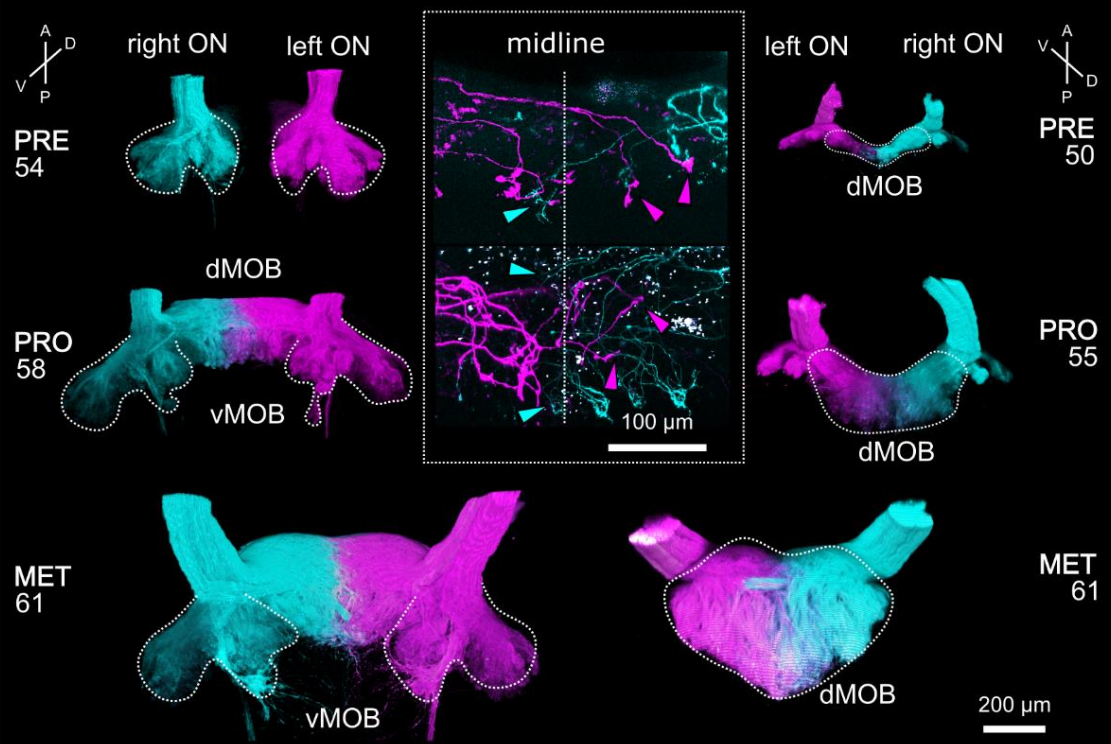
Distinct interhemispheric connectivity at the level of the olfactory bulb emerges during Xenopus laevis metamorphosis
Agrin/Lrp4 signal constrains MuSK activity during neuromuscular synapse development in appendicular muscle
Dichaete, a Sox2 homologue, prevents activation of cell death in multiple developmental contexts
Sperm cryopreservation impacts the early development of equine embryos by downregulating specific transcription factors
A single short reprogramming early in life improves fitness and increases lifespan in old age
| Morphogenesis & mechanics
Fly embryos from Perez-Vale, et al.
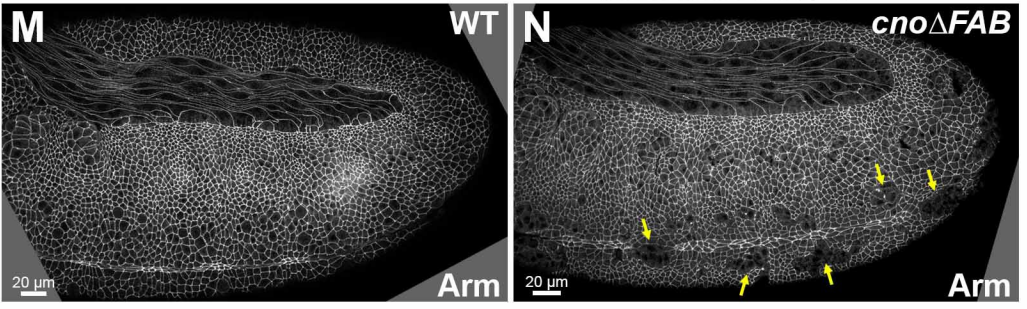
Multivalent interactions make adherens junction-cytoskeletal linkage robust during morphogenesis
Oligodendrocyte precursor cells guide the migration of cortical interneurons by unidirectional contact repulsion
Morphogenesis of a complex glial niche requires an interplay between cellular growth and fusion
Semaphorin3f as an intrinsic regulator of chamber-specific heart development
Frizzled3 inhibits Vangl2-Prickle3 association to establish planar cell polarity in the vertebrate neural plate
CD9 tetraspanins convey robustness to CXCR4b signalling during collective cell migration
Live imaging of delamination in Drosophila shows that epithelial cell motility and invasiveness are independently regulated
LMO7-dependent apical constriction requires the binding of the Myosin heavy chain
Syntaxin-1 is necessary for UNC5/Netrin-1-dependent macropinocytosis and chemorepulsion
Mouse skulls from Tsujikawa, et al.
Mechanical collaboration between the embryonic brain and the surrounding scalp tissues
Notch controls the cell cycle to define leader versus follower identities during collective cell migration
Actin-related protein 5 functions as a novel modulator of MyoD and MyoG in skeletal muscle and in rhabdomyosarcoma
Regulators of the secretory pathway have distinct inputs into single-celled branching morphogenesis and seamless tube formation in the Drosophila trachea
The zebrafish meiotic cohesion complex protein Smc1b is required for key events in meiotic prophase I
Cortical neurons from Creighton, et al.
Giant ankyrin-B mediates transduction of axon guidance and collateral branch pruning factor Sema 3A
Met is required for oligodendrocyte progenitor cell migration in Danio rerio
Fly ventral nerve cords from Howard, et al.
The Slit-binding Ig1 domain is required for multiple axon guidance activities of Drosophila Robo2
Kinesin-3 mediated delivery of presynaptic neurexin stabilizes growing dendritic spines and postsynaptic components in vivo
EVL and MIM/MTSS1 regulate actin cytoskeletal remodeling to promote dendritic filopodia in developing neurons
Knocking-out the human face genes TBX15 and PAX1 in mice alters facial and other physical morphology
Foregut organ progenitors and their niche display distinct viscoelastic properties in vivo during early morphogenesis stages
Dynein light chain-dependent dimerization of Egalitarian is essential for maintaining oocyte fate in Drosophila
SKAP2 as a new regulator of oligodendroglial migration and myelin sheath formation
Isoform-specific roles of the Drosophila filamin-type protein Jitterbug (Jbug) during development
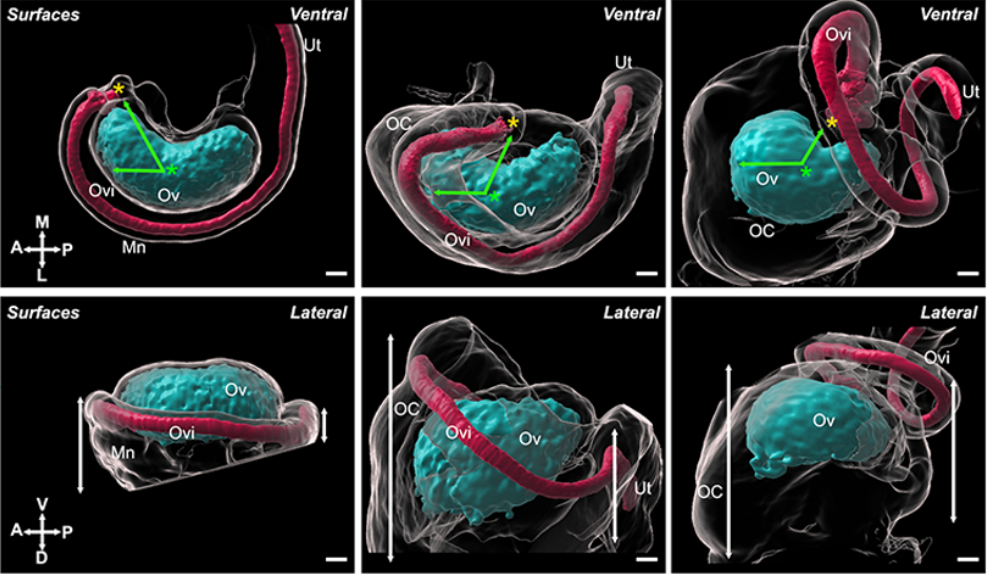
Mouse ovary reconstructions from McKay, et al.
Integration of mouse ovary morphogenesis with developmental dynamics of the oviduct, ovarian ligaments, and rete ovarii.
Transient nuclear deformation primes epigenetic state and promotes cell reprogramming
Mechanisms underlying microglial colonization of developing neural retina in zebrafish
| Genes & genomes
Mouse oocytes from Carpenter, et al.
CoREST has a conserved role in facilitating SPR-5/LSD1 maternal reprogramming of histone methylation
Epigenetic Inheritance is Gated by Naïve Pluripotency and Dppa2
Tiled C matric from Owens, et al.
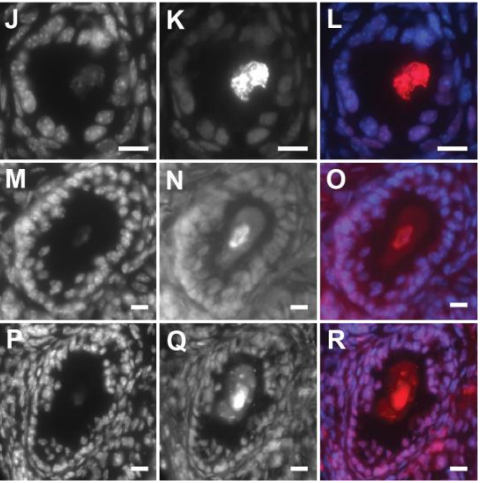
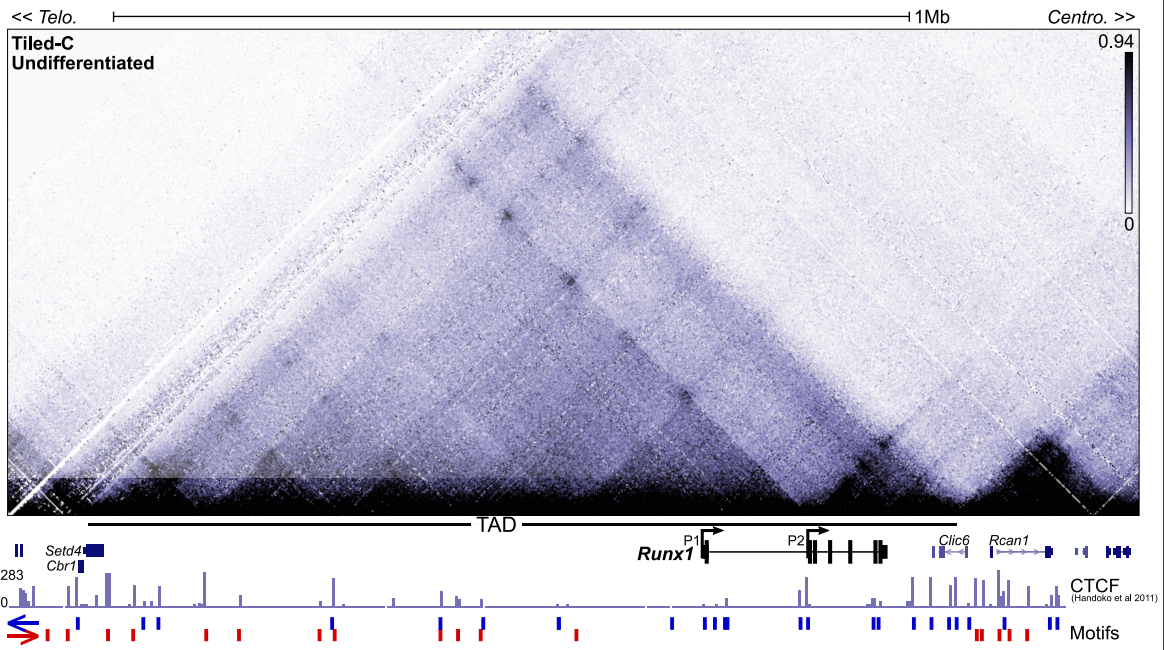
Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development
Foxd3 controls heterochromatin-mediated silencing of repeat elements in mouse embryonic stem cells and represses the 2-cell transcription program
In situ and transcriptomic identification of synapse-associated microglia in the developing zebrafish brain
Spatiotemporal specificity of correlated DNA methylation and gene expression pairs across different human tissues and stages of brain development
Quantitative comparison of in vitro and in vivo embryogenesis at a single cell resolution
Single-cell transcriptome analysis of the zebrafish embryonic trunk
Single-cell RNA-sequencing reveals thoracolumbar vertebra heterogeneity and rib-genesis in pigs
Molecular diversity and lineage commitment of human interneuron progenitors
Control of spinal motor neuron terminal differentiation through sustained Hoxc8 gene activity
Diverse mechanisms for epigenetic imprinting in mammals
Inferring kinetic parameters of oscillatory gene regulation from single cell time series data
Alternative somatic and germline gene-regulatory strategies during starvation-induced developmental arrest
Cell trajectory modeling identifies a primitive trophoblast state defined by BCAM enrichment
Fish flanks from Aman, et al.
Transcriptomic profiling of tissue environments critical for post-embryonic patterning and morphogenesis of zebrafish skin
The Hox gene Antennapedia regulates wing development through 20-hydroxyecdysone in insect
Dual origin and multiple neuropeptidergic trajectories of hypothalamic POMC progenitors revealed by developmental single-cell transcriptomics
Alternative Promoter use Governs the Expression of IgLON Cell Adhesion Molecules in Histogenetic Fields of the Embryonic Mouse Brain
delilah, prospero and D-Pax2 constitute a gene regulatory network essential for the development of functional proprioceptors
Single cell transcriptomics and developmental trajectories of murine cranial neural crest cell fate determination and cell cycle progression
Worm oocytes from Carlston, et al.
PQN-59 antagonizes microRNA-mediated repression and functions in stress granule formation during C. elegans development
BRD9-containing non-canonical BAF complexes safeguard cell identity and prevent reprogramming
Redundant enhancers in the iab-5 domain cooperatively activate Abd-B in the A5 and A6 abdominal segments of Drosophila
Genetic analysis of Caenorhabditis elegans pry-1/Axin suppressors identifies genes involved in reproductive structure development, stress response, and aging
Targeted DamID in C. elegans reveals a role for LIN-22 and NHR-25 in epidermal cell differentiation
Translation-dependent mRNA localization to Caenorhabditis elegans adherens junctions
Three-dimensional chromatin architecture of early-stage mouse embryos reconstructed via recurrence plots
| Stem cells, regeneration & disease modelling
Mouse embryos from Kinoshita, et al.
Disabling de novo DNA methylation in embryonic stem cells allows an illegitimate fate trajectory
RSL24D1 sustains steady-state ribosome biogenesis and pluripotency translational programs in embryonic stem cells
Neural stem cells alter nucleocytoplasmic partitioning and accumulate nuclear polyadenylated transcripts during quiescence
hPSC-derived endothelial progenitors from Gastfriend, et al.
Wnt signaling mediates acquisition of blood-brain barrier properties in naïve endothelium derived from human pluripotent stem cells
Definitive Hematopoietic Stem Cells Minimally Contribute to Embryonic Hematopoiesis
Embryonic stem cells from Votjek and Chambers
Resf1 supports embryonic stem cell self-renewal and effective germline entry
Translational specialization in pluripotency by RBPMS poises future lineage-decisions
Transcriptional Reprogramming of Skeletal Muscle Stem Cells by the Niche Environment
Craniofacial cartilage organoids from human embryonic stem cells via a neural crest cell intermediate
Self-organized yolk sac-like organoids allow for scalable generation of multipotent hematopoietic progenitor cells from human induced pluripotent stem cells
Wnt- and Glutamate-receptors orchestrate stem cell dynamics and asymmetric cell division
Soft limbal niche maintains stem cell compartmentalization and function through YAP
The RNA helicases DDX5 and DDX17 facilitate neural differentiation of human pluripotent stem cells NTERA2
Reprogrammed iBlastoids contain amnion-like cells but not trophectoderm
Hypertrophic Chondrocytes Serve as a Reservoir for Unique Marrow Associated Skeletal Stem and Progenitor Cells, Osteoblasts, and Adipocytes During Skeletal Development
Retinal organoids derived from rhesus macaque iPSCs undergo accelerated differentiation compared to human stem cells
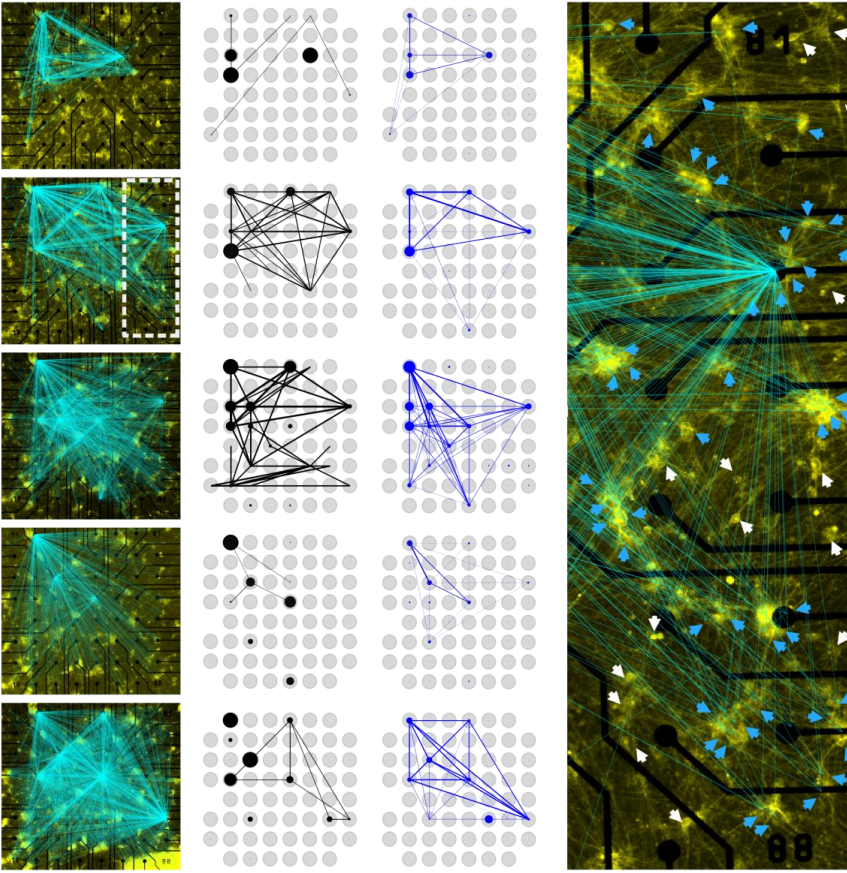
Functional connectivity in Schmieder, et al.
Tracking long-term functional connectivity maps in human stem-cell-derived neuronal networks by holographic-optogenetic stimulation
Edaravone activates the GDNF/RET neurotrophic signaling pathway and protects mRNA-induced motor neurons from iPS cells
Extracellular vesicles from neuronal cells promote neural induction of mESCs through cyclinD1
A secreted proteomic footprint for stem cell pluripotency
Derivation of ringed seal (Phoca hispida) induced multipotent stem cells
Stem cells commit to differentiation following multiple induction events in the Drosophila testis
Differentiation of cortical brain organoids and optic nerve-like structures from retinal confluent cultures of pluripotent stem cells
Novel epigenetic clock for fetal brain development predicts prenatal age for cellular stem cell models and derived neurons
Mesenchymal stem cell subpopulations and their heterogeneity of response to inductions revealed by single-cell RNA-seq
Generation of human chambered cardiac organoids from pluripotent stem cells for improved modelling of cardiovascular diseases
β-catenin perturbations control differentiation programs in mouse embryonic stem cells
Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects
Biomechanics and Myofibrillar Alignment Enhance Contractile Development and Reproducibility in Stem Cell Derived Cardiac Muscle
Edaravone activates the GDNF/RET neurotrophic signaling pathway and protects mRNA-induced motor neurons from iPS cells
ALKBH5 regulates somatic cell reprogramming in a phase specific manner
Regenerating fish fins in Heller, et al.
Characterization of mouse Bmp5 regulatory injury element in zebrafish wound models
Neuron-radial glial cell communication via BMP/Id1 signaling maintains the regenerative capacity of the adult zebrafish telencephalon
Hif1α is required for Wnt regulated gene expression during Xenopus tropicalis tail regeneration
Overexpression of Reticulon 3 enhances CNS axon regeneration and functional recovery after injury
Single-cell resolution of MET and EMT programs during zebrafish fin regeneration
Understanding the complexity of Epimorphic Regeneration in zebrafish: A Transcriptomic and Proteomic approach.
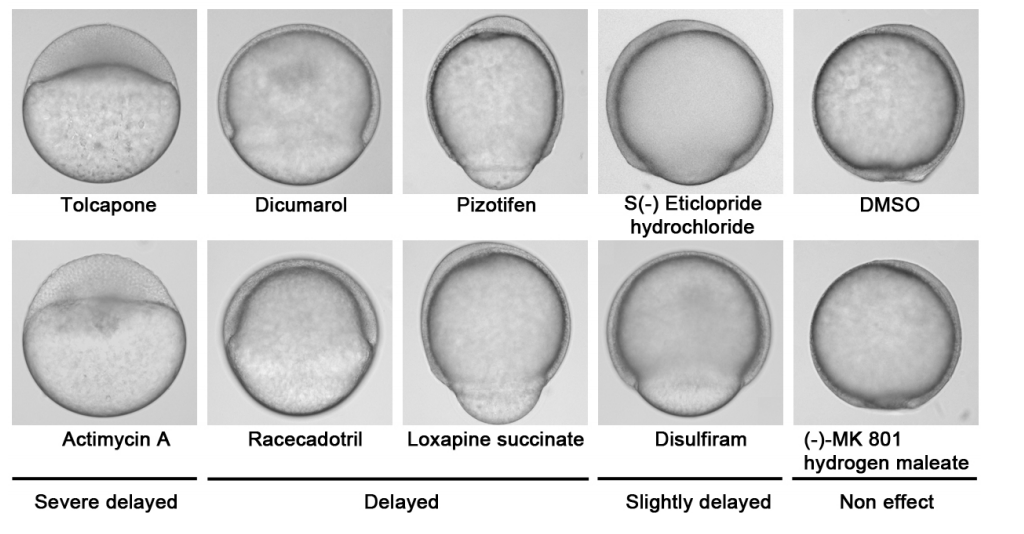
Nakayama, et al’s drug screen
A chemical screen based on an interruption of zebrafish gastrulation identifies the HTR2C inhibitor Pizotifen as a suppressor of EMT-mediated metastasis
iPSC modeling shows uncompensated mitochondrial mediated oxidative stress underlies early heart failure in hypoplastic left heart syndrome
Regulation of Neural Circuit Development by Cadherin-11 Provides Implications for Autism
mRNA-decapping associated DcpS enzyme controls critical steps of neuronal development
zmiz1a zebrafish mutants have defective erythropoiesis, altered expression of autophagy genes, and a deficient response to vitamin D
Correction of a pathogenic mutation in iPSCs derived from a patient with Christianson syndrome using CRISPR/Cas9 genome editing
Gains of 12p13.31 delay WNT-mediated initiation of hPSC differentiation and promote residual pluripotency in a cell cycle dependent manner
Maternal hyperglycemia impedes second heart field-derived cardiomyocyte differentiation to elevate the risk of congenital heart defects
In vitro models of the human esophagus reveal ancestrally diverse response to injury
The Drosophila orthologue of the primary ciliary dyskinesia-associated gene, DNAAF3, is required for axonemal dynein assembly
Forebrain Shh overexpression improves cognitive function in a Down syndrome mouse model and euploid littermates
SPT6 loss Permits the Transdifferentiation of Keratinocytes into an Intestinal Fate that Recapitulates Barrett’s Metaplasia
| Plant development
Genome-Wide High Resolution Expression Map and Functions of Key Cell Fate Determinants Reveal the Dynamics of Crown Root Development in Rice
Tissue-specific transcriptomics reveal functional differences in maize floral development
Rice tissues from Rong, et al.
Cytokinin oxidase/dehydrogenase family genes play important roles in the growth and development of rice
KAI2 regulates seedling development by mediating light-induced remodelling of auxin transport
Phototropin-mediated perception of light direction in Arabidopsis leaves regulates blade flattening
Misregulation of MYB16 causes stomatal cluster formation by disrupting polarity in asymmetric cell division
Arabidopsis cotyledons from mHan, et al.
Deceleration of cell cycle underpins a switch from proliferative- to terminal division in plant stomatal lineage
Microtubules Promote the Non-cell Autonomy of MicroRNAs by Inhibiting their Cytoplasmic Loading into ARGONAUTE1 in Arabidopsis
The ALOG family members OsG1L1 and OsG1L2 regulate inflorescence branching in rice
A DUF1068 protein acts as a pectin biosynthesis scaffold and maintains Golgi morphology and cell adhesion in Arabidopsis
DELAY OF GERMINATION 6, encoding the ANAC060 transcription factor, inhibits seed dormancy
The role of AUX1 during lateral root development in the domestication of the model C4 grass Setaria italica
Biphasic Control of Cell Expansion by Auxin Coordinates Etiolated Seedling Development
Functional characterization of TANGLED1 interaction with PHRAGMOPLAST ORIENTING KINESIN1 during mitosis in Arabidopsis
Actin isovariant ACT7 regulates root meristem development in Arabidopsis through modulating auxin and ethylene responses
GIGANTEA gene expression influence leaf senescence in Populus in two different ways
Early flowering in oilseed-type Brassica rapa plants results from nonsense-mediated mRNA decay (NMD) of BrFLC2
GA-mediated spatial control of cell division expounds the leaf size variation between cultivated and wild rice
Arabidopsis plants from Dong, et al.
AGL16 regulates genome-wide gene expression and flowering time with partial dependency on SOC1 in Arabidopsis
PIF7 controls leaf cell proliferation through an AN3 substitution-repression mechanism
Dual functions of ZmGI1 in the photoperiodic flowering pathway and salt stress responses in maize
OsbZIP47 an integrator for meristem regulators during rice plant growth and development
Transcriptome-wide identification and expression profiling of the ERF gene family suggest roles as transcriptional activators and repressors of fruit ripening in durian
Combined fluorescent seed selection and multiplex CRISPR/Cas9 assembly for fast generation of multiple Arabidopsis mutants
| Evo-devo
Single cell RNA sequencing of the Strongylocentrotus purpuratus larva reveals the blueprint of major cell types and nervous system of a non-chordate deuterostome
ERK1/2 is an ancestral organising signal in spiral cleavage
Species-specific deployment of Runx2 isoforms and differential regulation of target genes during avian jaw development and evolution
BMP signaling underlies the craniofacial heterochrony in phyllostomid bats, a hyperdiverse mammal group
Capitella embryos from webster, et al.
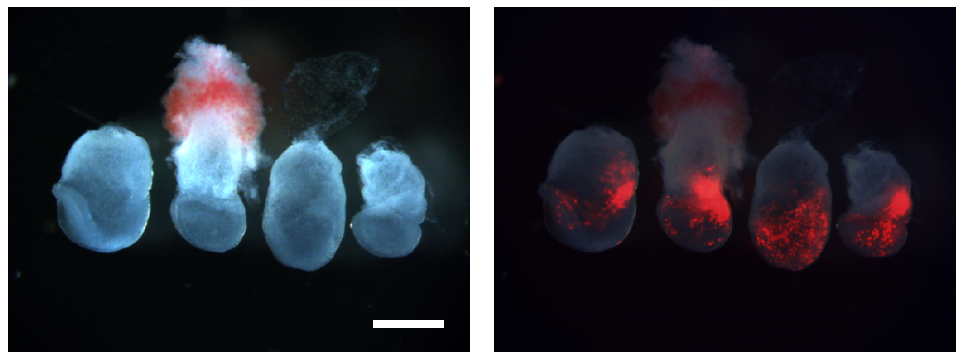
Role of BMP signaling during early development of the annelid Capitella teleta
Single gene initiates evolution of epithelial architecture and function
Pigment pattern morphospace of Danio fishes: evolutionary diversification and mutational effects
Revealing conserved mechanisms of neurodegeneration in a colonial chordate
Species-specific developmental timing dictates expansion of the avian wing skeletal pattern
Loricarioid catfish evolved skin denticles that recapitulate teeth at the structural, developmental, and genetic levels
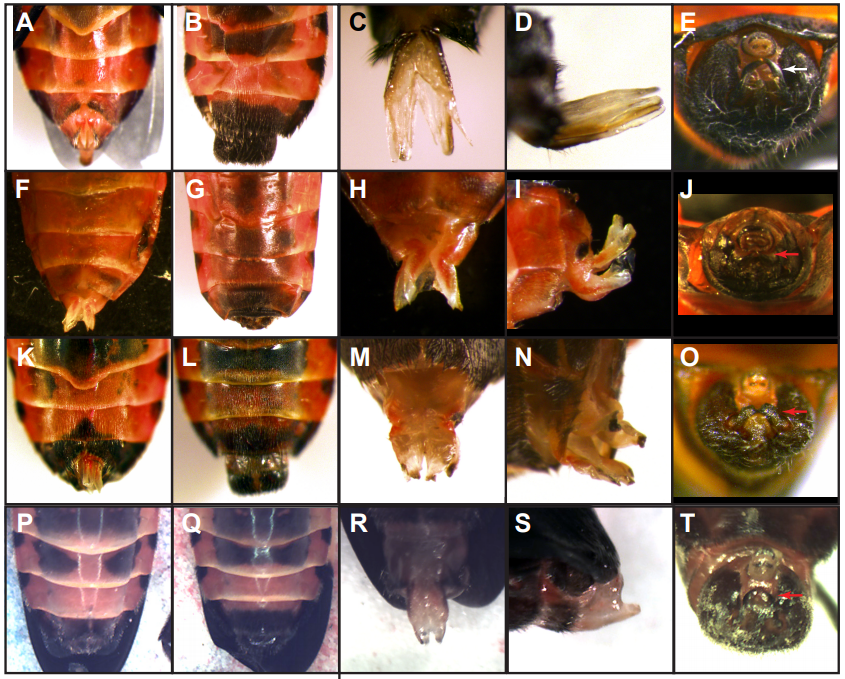
Oncopeltus abdomens from Just, et al.
Distinct developmental mechanisms influence sexual dimorphisms in the milkweed bug Oncopeltus fasciatus
A 3D molecular map of the cavefish neural plate illuminates eyefield organization and its borders in vertebrates
Evolved Bmp6 enhancer alleles drive spatial shifts in gene expression during tooth development in sticklebacks
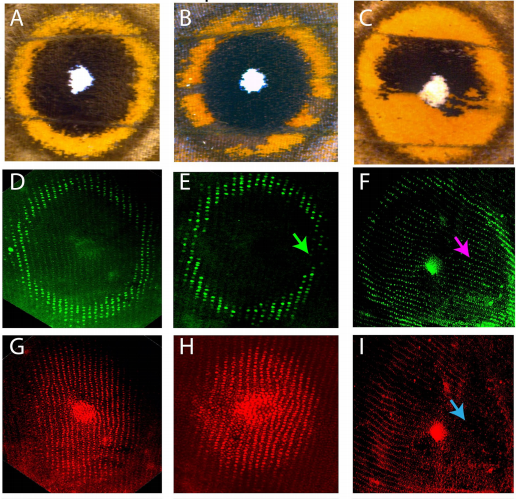
Wing spots from Banerjee, et al.
optix is involved in eyespot development via a possible positional information mechanism
Establishment of CRISPR/Cas9-based knock-in in a hemimetabolous insect: targeted gene tagging in the cricket Gryllus bimaculatus
The transcriptome of Schistosoma mansoni developing eggs reveals key mediators in pathogenesis and life cycle propagation
Social selection within aggregative multicellular development drives morphological evolution
Cell Biology
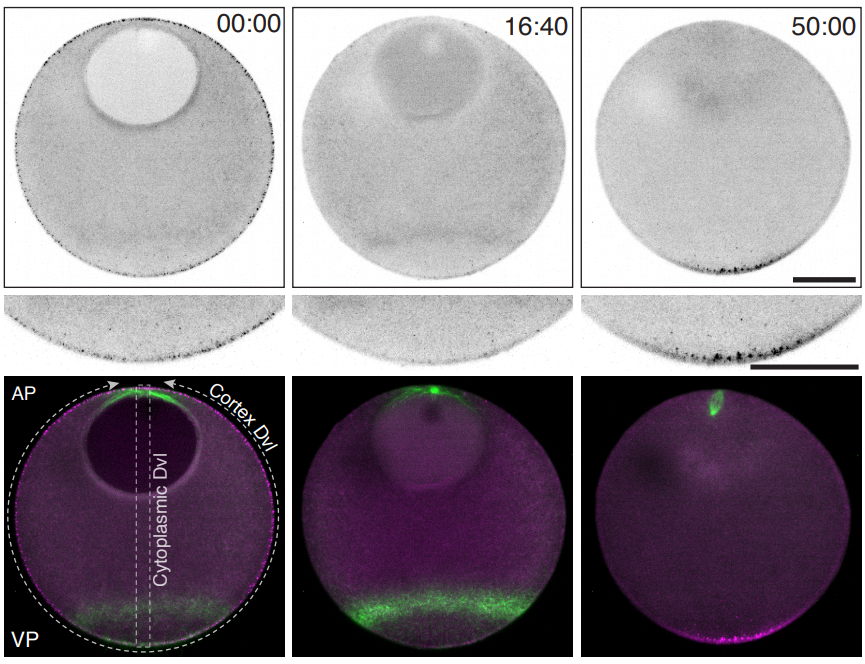
Sea star oocytes from Swartz, et al.
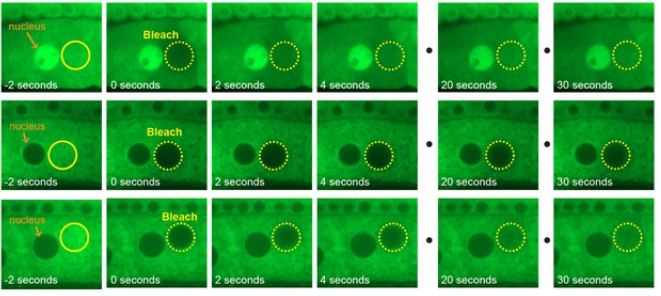
Polarized Dishevelled dissolution and condensation drives embryonic axis specification in oocytes
Combined effect of cell geometry and polarity domains determines the orientation of unequal division
Building the cytokinetic contractile ring in an early embryo: initiation as clusters of myosin II, anillin and septin, and visualization of a septin filament network
Septin function tunes lipid kinase activity and phosphatidylinositol 4,5 bisphosphate turnover during G-protein coupled PLC signaling in vivo
Mad1’s ability to interact with Mad2 is essential to regulate and monitor meiotic synapsis in C. elegans
xbx-4, a homolog of the Joubert syndrome gene FAM149B1, acts via the CCRK and MAK kinase cascade to regulate cilia morphology
Septins and a formin have distinct functions in anaphase chiral cortical rotation in the C. elegans zygote
Tmem138, a photoreceptor connecting cilium (CC) protein, is required for rhodopsin transport across the cilium and outer segment (OS) biogenesis
Nance-Horan Syndrome-like 1 protein negatively regulates Scar/WAVE-Arp2/3 activity and inhibits lamellipodia stability and cell migration
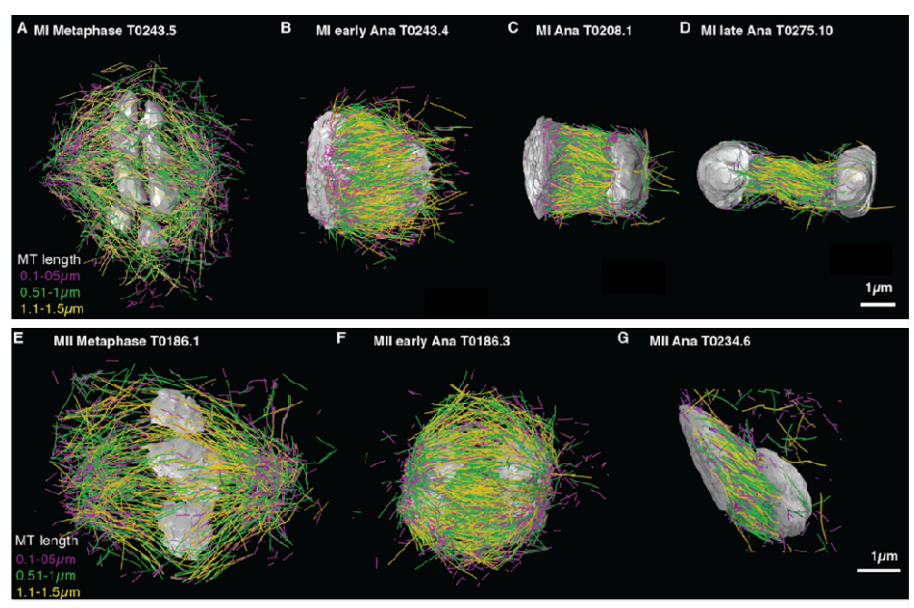
Worm meiosis from Lantzsch, et al.
Microtubule re-organization during female meiosis in C. elegans
Scaling of cellular proteome with ploidy
GIV/Girdin, a Non-receptor Modulator for Gαi/s, Regulates Spatiotemporal Signaling during Sperm Capacitation and is Required for Male Fertility
WASP integrates substrate topology and cell polarity to guide neutrophil migration
Paternal chromosome elimination and X non-disjunction on asymmetric spindles in Sciara male meiosis
Mechanosensitive calcium signaling in response to cell shape changes promotes epithelial tight junction remodeling by activating RhoA
WASP integrates substrate topology and cell polarity to guide neutrophil migration
Modelling
A coarse-grained approach to model the dynamics of the actomyosin cortex
A mathematical framework for evo-devo dynamics
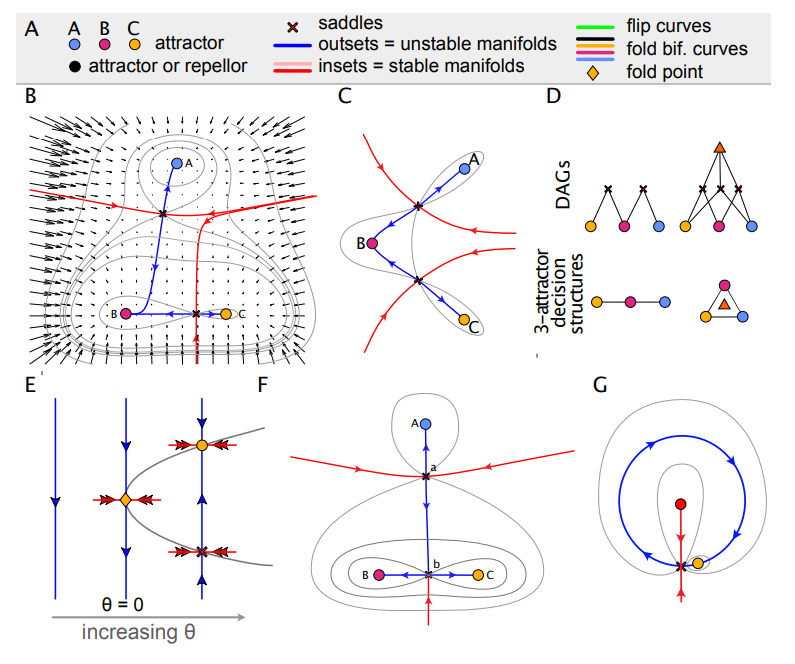
Morse-Smale systems in Rand, et al.
Geometry of Gene Regulatory Dynamics
Spontaneous cell internalization of a spatially-confined proliferating blastomere: A mechanical interpretation on worm gastrulation
Novel Generic Models for Differentiating Stem Cells Reveal Oscillatory Mechanisms
Hybrid reaction-diffusion and clock-and-wavefront model for the arrest of oscillations in the somitogenesis segmentation clock
A mathematical model of endothelial progenitor cell cluster formation during the early stages of vasculogenesis
Improving the understanding of cytoneme-mediated morphogen gradients by in silico modeling
Revealing cell-fate bifurcations from transcriptomic trajectories of hematopoiesis
Self-organization principles of cell cycles and gene expressions in the development of cell populations
Generation of fate patterns via intercellular forces
Travelling wave analysis of cellular invasion into surrounding tissues
Reviews
Then There were Plenty – Ring Meristems Giving Rise to Many Stamen Whorls
Modulation of Organogenesis and Somatic Embryogenesis by Ethylene: An Overview
Thermogenic Fat: Development, Physiological Function, and Therapeutic Potential
A Drosophila Toolkit for Imaging of HA-tagged Proteins Unveiled a Block in Autophagy Flux in the Last Instar Larval Fat Body
Tracking organoids in de Medeiros, et al.
Multiscale light-sheet organoid imaging framework
Whole-ExM: Expansion microscopy imaging of all anatomical structures of whole larval zebrafish
Learning dynamics by computational integration of single cell genomic and lineage information
Tools for efficient analysis of neurons in a 3D reference atlas of whole mouse spinal cord
A Cre-dependent massively parallel reporter assay allows for cell-type specific assessment of the functional effects of genetic variants in vivo
Evaluation of CRISPR gene-editing tools in zebrafish
Hnf1b-CreER, a model used for fate mapping pancreatic lineages, achieves efficient Cre-mediated recombination in duct and islet δ cells
Leveraging single-cell ATAC-seq to identify disease-critical fetal and adult brain cell types
The FusX TALE Base Editor (FusXTBE) for rapid mitochondrial DNA programming of human cells in vitro and zebrafish disease models in vivo
The adenoviral E1B-55k protein present in HEK293 cells mediates abnormal accumulation of key WNT signaling proteins in large cytoplasmic aggregates
Confounds of using the unc-58 selection marker highlights the importance of genotyping co-CRISPR genes
High resolution, serial imaging of early mouse and human liver bud morphogenesis in three dimensions
Origami: Single-cell oriented 3D shape dynamics of folding epithelia from fluorescence microscopy images
Multifocal imaging for precise, label-free tracking of fast biological processes in 3D
Live 3D imaging and mapping of shear stresses within tissues using incompressible elastic beads
Improved methods for protein and single-molecule RNA detection in C. elegans embryos
Rapid generation of conditional knockout mice using the CRISPR-CAS9 system and electroporation for neuroscience research
Instant three color multi-plane fluorescence microscopy
Label-free imaging flow cytometry: analysis and sorting of enzymatically dissociated tissues
Genome-wide screening in human kidney organoids identifies novel aspects of nephrogenesis
An optimized protocol for neuronal assay for transposase-accessible chromatin by sequencing (ATAC-seq) library preparation using Drosophila melanogaster
Multi-view confocal microscopy enables multiple organ and whole organism live-imaging
AZBA: A 3D Adult Zebrafish Brain Atlas for the Digital Age
Minian: An open-source miniscope analysis pipeline
Crossbill: an open access single objective light-sheet microscopy platform
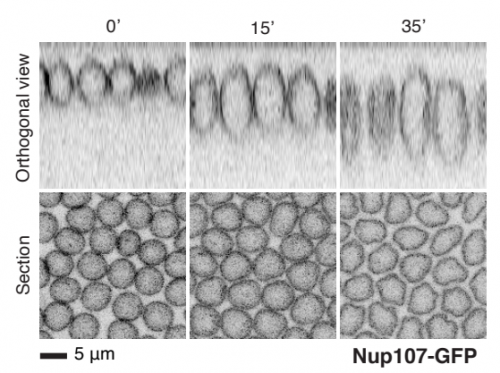
Fly nuclear pores from Cheng, et al.
A method for single molecule localization microscopy of tissues reveals nonrandom distribution of nuclear pores in Drosophila
Microfabricated disk technology: rapid scale up in midbrain organoid generation
maplet: An extensible R toolbox for modular and reproducible omics pipelines
A Universal Approach to Analyzing Transmission Electron Microscopy with ImageJ
A new pipeline to automatically segment and semi-automatically measure bone length on 3D models obtained by Computed Tomography
Label-free three-dimensional analyses of live cells with deep-learning-based segmentation exploiting refractive index distributions
Tracking worm meiosis in Cahoon and Libuda.
Conditional immobilization for live imaging C. elegans using auxin-dependent protein depletion
Research practice & education
“How do we do this at a distance?!” A descriptive study of remote undergraduate research programs during COVID-19
1 votes)



 (No Ratings Yet)
(No Ratings Yet)