From mysterious cysts to CSF-in-a-dish
Posted by Laura Pellegrini, on 21 September 2020
Our brain is immersed in a clear, colourless, nutrient-rich fluid called the cerebrospinal fluid (CSF), which provides mechanical support to the brain and helps to circulate important molecules for brain development and function. Within the interconnected cavities of our brain, the CSF flows in and out constantly. The CSF is actively produced by the choroid plexus (ChP), a highly secretory tissue present at different locations inside the brain cavities.
The ChP-CSF system plays a number of central roles in our brain, such as actively secreting and transporting hormones and nutrients to different brain regions and maintaining the physiological balance of ions and salts. In addition, the ChP is implicated in circadian cycle regulation, influencing the master clock regulation in the brain, and during sleep, CSF flow has been proposed to remove toxic protein aggregates and waste from the brain. Another key function of the ChP is the formation of a selectively permeable barrier, the blood-CSF-barrier, which separates the CSF from the blood, protecting the brain from toxic substances and certain drugs, similar to the blood-brain-barrier.
Despite all these important functions, the ChP and the CSF have long been ignored or dismissed simply as a waste disposal system of the brain, therefore for a long time little was known about these fascinating components of our brain. We now understand a lot more of the development and function of this tissue thanks to research on animal models such as mice, however, the study of human ChP and CSF is still complicated by technical and ethical challenges to obtain biopsies of ChP, particularly at early developmental stages. Studying human CSF is also particularly challenging due to the fact that extraction of this fluid requires painful lumbar punctures which run the risk of blood contamination.
In Madeline Lancaster’s lab at the MRC-LMB in Cambridge, we routinely generate cerebral organoids from human pluripotent stem cells. Cerebral organoids develop neural tissue which expands and matures forming structures that resemble early developmental stages of our brain’s dorsal cortex. Organoids are placed in a supporting media that contains only the essential nutrients and key ingredients that help and sustain brain development. This essential media allows for the spontaneous development of the brain tissue as it would normally develop in vivo, so sometimes we observe the generation of other brain identities other than the desired cerebral cortex. It was in this way that we initially observed the presence of ChP-like tissue adjacent to part of the cerebral cortex. It was this initial observation that prompted us to reproducibly generate ChP tissue. In our recent study entitled “Human CNS barrier-forming organoids with cerebrospinal fluid production” (Pellegrini et al., 2020), we developed a new in vitro model to study early development of human ChP in a dish.
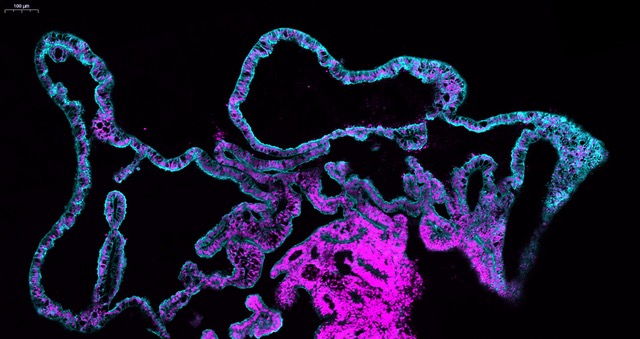
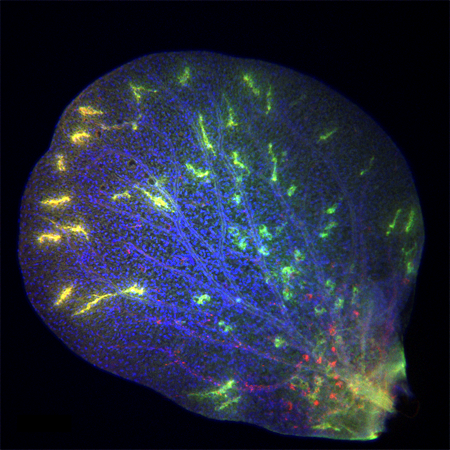
We started using patterning molecules secreted in vivo and previously shown to model ChP development in vitro (Sakaguchi et al., 2015; Watanabe et al., 2012). By adding these molecules at certain timepoint and for a certain duration, the organoids became entirely enriched in ChP tissue. To our surprise we noticed that these organoids would develop various-shaped, clear looking cysts that appeared completely isolated from the pink media surrounding the organoid. Initially, because of the unknown identity of these mysterious cysts, many of these organoids were discarded or considered “failed organoids”. One day, instead of throwing these organoids away we decided to look more carefully at them. By sectioning and staining the tissue with specific markers we came to understand that indeed the cyst was entirely surrounded by ChP.
The first question we had was: are these fluid compartments filled with human CSF? We were able to extract several millilitres of this clear fluid from different organoids. We then decided to perform mass spectrometry analysis to investigate the overall protein composition of the fluid. Together with the fluid extracted from organoids, we also analysed the media as a “negative control” and human in vivo CSF from healthy donors as a “positive control”. The results were quite surprising. The first observation was that this intriguing organoid fluid was not a mere ultrafiltrate of the surrounding media, but a fluid that contained proteins secreted de novo by the ChP. Second, looking at the proteins secreted de novo by the organoid we could find several human CSF proteins, including specific signalling molecules secreted during development, such as Insulin Growth Factor 2 (IGF2), and disease-relevant biomarker such as Serpin Family F Member 1 (SERPINF1) and clusterin (CLU) were indeed present in the organoid fluid, but not in the media. Comparing CSF extracted from organoids at different stages, we could notice a difference in the protein composition, with an enrichment in extracellular matrix proteins at early stages, compared to later stages in which we detected several apolipoproteins involved in lipid metabolism. When we compared our dataset with published proteomic databases of CSF in vivo, we realised that these differences in CSF composition reflected what is reported in embryonic CSF compared to older post-natal CSF stages, matching different stages of maturation.
Another interesting observation is that our organoid-generated CSF lacked blood proteins, such as haemoglobin, which were instead detected in CSF from humans. This is because organoids lack vasculature. We suspect that the presence of these proteins in human CSF is due to contamination from the lumbar punctures used to extract the fluid. Lacking these potential contaminants, the organoid-CSF represents a “cleaner” version of the ChP secretome. These results meant that we were able to generate a close to authentic CSF in a dish, from human cells. Compared to the small amount of CSF one can obtain from mouse embryos, we could extract several millilitres and we could look at the composition of this fluid without any other contaminants.
As mentioned above the ChP forms the blood-CSF-barrier, which regulates the exchange of nutrients and signals from the blood to the CSF, blocking entry of different compounds such as therapeutics. This brain barrier is less studied than the blood-brain-barrier, but it could provide an alternative way to access the brain if appropriately targeted. We therefore wondered: does the ChP in these organoids form a selectively permeable barrier?
Initially, we had a hint that the answer was yes because the liquid inside these cysts is clear, meaning that even the small molecule of phenol red, usually present in our media, is not able to freely cross the barrier. Since we could extract significant amount of fluid from these organoids, the first experiment was simply to add fluorescently labelled molecules of different sizes to the media and observe, after 2 hours of incubation, whether we could detect any fluorescence in the inside of the fluid-filled organoid compartment. Even the smallest of these molecules tested was undetectable inside of the organoid, indicating that the ChP in vitro was indeed forming a barrier. We then wanted to know whether the ChP forming the barrier could select certain molecules instead of others.
While having a coffee with brilliant chemist and friend Dr. Claudia Bonfio, who also works at the MRC-LMB in the completely unrelated field of prebiotic chemistry, we discussed possible ways to detect small molecules in their native state, in a system like the ChP organoids. This unusual meeting between chemistry and biology resulted in the idea of using Nuclear Magnetic Resonance (NMR). Using this technique, we were able to demonstrate the selective permeability of the ChP barrier by challenging the system with two highly similar small molecules with opposite permeabilities such as L-dopa and dopamine. We indeed observed that only L-dopa, and not dopamine, was able to cross the ChP barrier, exactly recapitulating the selectivity observed in vivo. This initial proof of principle experiment was fundamental for us to trust the system and then challenge it with other drugs.
With this experimental approach we were able to show that organoids can qualitatively but also quantitatively predict the permeability of new drugs. When we tested the new drug Sephin 1, a compound that prevents protein misfolding that has recently entered clinical trials, we noticed that the permeability data were slightly lower in the organoids compared to the available data in animals, in this case mice. This indicates that the system could recapitulate permeability of drugs in human, but the levels of transporters expressed in the ChP can be different in other species, reflecting differences in permeability. Finally, we decided to use our method to reveal why the drug BIA 10-2474, which failed clinical trials for multiple conditions including chronic pain and multiple sclerosis, was toxic only in humans. We indeed observed an unusual accumulation of this compound inside the organoid fluid. This observation was consistent with the hypothesis of an accumulation in the CSF of these patients. Together with other toxic effects reported for this drug, the use of ChP organoids could have perhaps have raised some flags during preclinical testing. We think that ChP organoids potentially represent a preclinical in vitro human model to validate safe entry of therapeutics to the brain, with the potential to reduce the number of drugs that fail during clinical trials.
Finally, we were curious to explore in more detail the cellular architecture of the choroid plexus in order to better characterise the different cell populations present in the system. We took advantage of this isolated nature of ChP organoids, not attached to other tissues, to identify cell populations unique to this region of the brain. We looked closely at the ChP and by merging our single cell and proteomic data we were able to learn which specific cell types secrete the biomarkers detected in the organoid CSF fluid. In addition, we identified these obscure “dark” and “light” cells that were first reported in electron micrographs from the ‘70s, but we were also able to discover a new cell type, previously unidentified in the ChP: the myoepithelial cells. These cells have been reported in other secretory tissues such as salivary and mammary glands, and they are enriched in contractile fibres which could potentially help to “squeeze” the secretory epithelial cells to help CSF production.
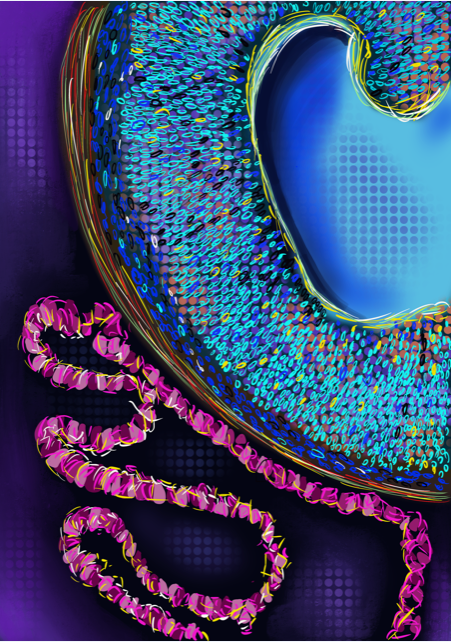
In summary, we learnt that ChP organoids could secrete CSF and form a selectively permeable barrier that can be used to select drugs in a human-specific system in vitro. We also learnt that if you notice something unusual in your experiments, don’t just throw it away!
References


 (4 votes)
(4 votes) (No Ratings Yet)
(No Ratings Yet)
 (11 votes)
(11 votes)