A day in the life of a colonial tunicate laboratory
Posted by Stefania Gutierrez, on 28 August 2018
Have you heard of an animal that can lose most of its body tissues and the remnant tissues aggregate to regenerate the lost parts and recovery its original form?
Do you know an animal that can quickly colonize marine surfaces by asexual reproduction, just like weed would in terrestrial environments ?
Do you know an animal that can disperse to new locations when small portions of its body separate and move away from the rest?
These descriptions may remind you of characters from science fiction stories, but these are real characteristics of colonial animals. Animals colonies are composed of discrete multicellular units (e.g zooids, polyps), that are physiologically interconnected and undergo clonal replication maintaining an identical genotype throughout all of their components (Hughes, 1989; Jackson & Coates, 1986) . This includes a number of marine and freshwater animals, such as many corals and hydroids, bryozoans, some hemichordates and some ascidians (a group within the subphylum Tunicata). One of the main projects in the laboratory is to study the evolution of colonial life strategy in ascidians, in which coloniality evolve several times.
I am a PhD student in Prof. Federico Brown’s laboratory (http://zoologia.ib.usp.br/evodevo2/) in the Department of Zoology at the University of São Paulo. We are located in a subtropical area in the south of Brazil, in a huge metropolis 100 km away from the ocean. In fact, our lab is located next to a small forest reserve on campus that is reminiscent of the Mata Atlantica Forest (a hotspot of biodiversity), from the laboratory’s windows, we can see a great diversity of birds, insects, spiders and sometimes even monkeys (Fig.1).
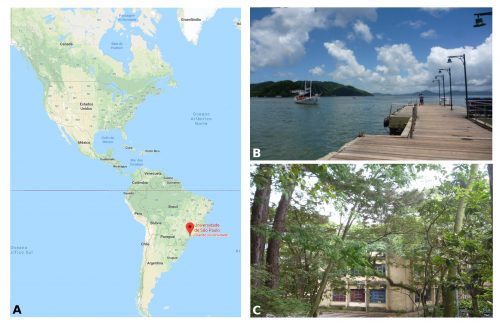
Our work in the laboratory begins with the collection of tunicates from the ocean. We travel by car to close coastal cities, including São Sebastião (4h away) and Santos (1h away) (Fig. 1B). We collect the tunicates from pilings and floats in the port marinas. The colonial species are carefully collected and attached to glass slides using thread (Fig. 2A). The slides are placed in slide boxes, with open windows on both sides to allow water circulation, and the boxes are hung to the dock with ropes. After two weeks, the colonies grow to cover the slides. Therefore attached tunicates are cleaned, transported to the aquarium system, and are ready for use in experiments (Fig. 2).

For my PhD project, I am working with the genus Symplegma, a member of the family Styelidae, in which coloniality arose at least twice from solitary ancestors. Symplegma is the sister genus to the botryllids (Botryllus and Botrylloides), a group of highly integrated colonial species that undergo weekly cycles of asexual development (Brown et al., 2009). Symplegma has less integrated colonies and characteristics more similar to the solitary species. Thus it is an interesting genus to study the evolution of coloniality.
Symplegma, as other colonial ascidians, has internal fertilization and brooding, with embryos that are incubated for several days to weeks before the tadpole larvae are released. The tadpole larva have a brief period of swimming and settle rapidly (Fig. 3A). During settlement, the larvae undergo metamorphosis, in which most of the tail structures are resorbed and the mouth rotates close to 90º to the dorsal side, changing from a tailed swimming larva into a sessile filter feeder (Fig. 3).
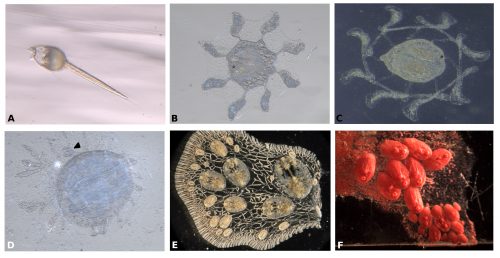
During metamorphosis, the ampullae extending a symmetrical pattern around the first zooid (Fig. 3B). The ampullae are peripheral pouch-like structures of the blood vessel system, essential for communication of the colony with its environment. Then ampullae continue to extend and, interconnect to form the primary net of blood vessels, in which the blood circulate constantly (Fig. 3C). Then this system of vessels grows forming more vessels that connect the zooid and the first bud (Fig. 3D).
Inside the vessels, specialized blood cells circulate, constantly coordinating biological processes between the zooids of the colony (Video 1).
For example, the circulatory cells are key to the clonal formation of new buds and the coordination of death of old zooids. New buds can be formed in two locations, either at the lateral epithelium of the adult zooid or along blood vessels that are far from adult zooids (Fig. 3E-F).
While some colonial styelids have highly integrated colonies with coordinated cycles of degeneration and regeneration of zooids, Symplegma is much less integrated and coordinated. We are interested in how developmental and regenerative processes differ between species with different degrees of coordination and integration. For example, we conduct experiments to observe whole body regeneration by removing all the zooids and buds retaining only the blood vessels of the original colony. Immediately after surgery, the blood coagulates and circulation in the remnant vessels stops. Twelve hours after the surgery, blood circulation is restored without any zooids or heart to pump the blood. Presumably, this circulation is caused by vessel contractions (Video 2).
Next, the remaining vascular tissue aggregates and form a mass, in which new zooids arise. Ten days after surgery a complete functional colony has regenerated (Fig.4).
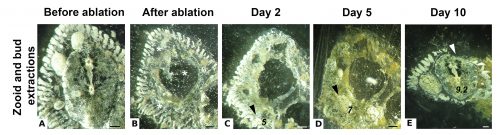
These results suggest that the vascular tissue has the capacity to rearrange itself and regenerate new zooids. Our results show that Symplegma colonies act like self-regulating systems that have the ability to rearrange its components (blood vessels and blood cells) after perturbations to regenerate damaged or lost parts. Due to the presence of replaceable zooids, the colonial life history allows for the recovery of lost parts by regeneration, fast colonization of marine substrates, and high survival rates after predation or weather-related disturbances. These colonial animals are like superorganisms!! They show amazing developmental mechanisms linked to coloniality.
If you would like any more information about their life history and how to work on them, just ask us in the comments or via email [as.gutierrez57@ib.usp.br].
References
-Brown, F. D., Tiozzo, S., Roux, M. M., Ishizuka, K., Swalla, B. J., & De Tomaso, A. W. (2009). Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development (Cambridge, England), 136(20), 3485–3494.https://doi.org/10.1242/dev.037754
-Gutierrez, S., & Brown, F. D. (2017). Vascular budding in Symplegma brakenhielmi and the evolution of coloniality in styelid ascidians. Developmental Biology, 423(2). https://doi.org/10.1016/j.ydbio.2017.01.012
-Hughes, R. (1989). A Functional Biology of Clonal Animals. New York: Chapman and Hall.
-Jackson, J. B. C., & Coates, a. G. (1986). Life Cycles and Evolution of Clonal (Modular) Animals. Philosophical Transactions of the Royal Society B: Biological Sciences, 313(1159), 7–22. https://doi.org/10.1098/rstb.1986.0022


 (4 votes)
(4 votes) (No Ratings Yet)
(No Ratings Yet)

 (4 votes)
(4 votes)