In Development this week (Vol. 144, Issue 20)
Posted by the Node, on 17 October 2017
Here are the highlights from the current issue of Development:
Sequencing sheds light on human spermatogenesis
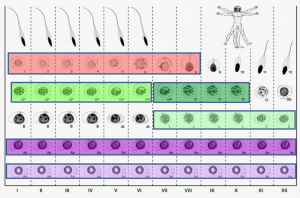 On p. 3659, Sjoerd Repping and colleagues combine laser-capture microdissection and next-generation RNA sequencing techniques to provide the first detailed analysis of the gene expression patterns of spermatogenic cells isolated from human tissue. They profile the transcriptomes of six functionally distinct classes of germ cells from the human testis, which represent successive steps in the process of spermatogenesis. They find that over 4000 genes are dynamically transcribed throughout this process, including a significant number of post-transcriptional regulators such as RNA-binding proteins and long non-coding RNAs. These data suggest that post-transcriptional regulatory mechanisms are important for the transition of germ cells from a precursor state into differentiated sperm. This work provides valuable insight into how the process of sperm production differs between humans and other species at a transcriptional level, and should serve as an important resource for identifying genes implicated in male infertility.
On p. 3659, Sjoerd Repping and colleagues combine laser-capture microdissection and next-generation RNA sequencing techniques to provide the first detailed analysis of the gene expression patterns of spermatogenic cells isolated from human tissue. They profile the transcriptomes of six functionally distinct classes of germ cells from the human testis, which represent successive steps in the process of spermatogenesis. They find that over 4000 genes are dynamically transcribed throughout this process, including a significant number of post-transcriptional regulators such as RNA-binding proteins and long non-coding RNAs. These data suggest that post-transcriptional regulatory mechanisms are important for the transition of germ cells from a precursor state into differentiated sperm. This work provides valuable insight into how the process of sperm production differs between humans and other species at a transcriptional level, and should serve as an important resource for identifying genes implicated in male infertility.miR-290: the making of placental mammals?
MicroRNAs play a variety of roles during development, primarily by regulating gene expression through repression of target mRNAs. One cluster, the miR-290 cluster, is specific to placental mammals, and has, in the past, been linked to pluripotency maintenance in the early mammalian embryo. 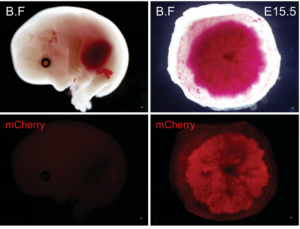 On p. 3731, Robert Blelloch and colleagues now uncover a role for the miR-290 cluster in placental development. Using transgenic mouse lines, they find that the miR-290 cluster is highly expressed during early mouse embryogenesis and becomes restricted to the trophoblast from gastrulation up until birth. When the miR-290 cluster is deleted, mRNAs targeted by this cluster become dysregulated. Interestingly, this deletion also causes several placental defects, including a reduction in trophoblast proliferation, reduced giant cell endoreduplication, and disruption of the vasculature of the placental labyrinth. Ultimately, this results in the development of a placenta that is reduced in size, and defective in passive diffusion of nutrients from the mother to the foetus, leading to late embryonic lethality. These results suggest that microRNAs within the miR-290 cluster are responsible for regulating the gene network important for placental growth and development in mice, and may provide insight into the evolution of eutherian mammals.
On p. 3731, Robert Blelloch and colleagues now uncover a role for the miR-290 cluster in placental development. Using transgenic mouse lines, they find that the miR-290 cluster is highly expressed during early mouse embryogenesis and becomes restricted to the trophoblast from gastrulation up until birth. When the miR-290 cluster is deleted, mRNAs targeted by this cluster become dysregulated. Interestingly, this deletion also causes several placental defects, including a reduction in trophoblast proliferation, reduced giant cell endoreduplication, and disruption of the vasculature of the placental labyrinth. Ultimately, this results in the development of a placenta that is reduced in size, and defective in passive diffusion of nutrients from the mother to the foetus, leading to late embryonic lethality. These results suggest that microRNAs within the miR-290 cluster are responsible for regulating the gene network important for placental growth and development in mice, and may provide insight into the evolution of eutherian mammals.
Semaphorin marks the spot in the pancreas
Islets are clusters of endocrine cells in the pancreas that contain specialised cell types responsible for the secretion of hormones such as insulin and glucagon. During the development of the pancreas, progenitors of islet cells delaminate from the embryonic ductal epithelium then migrate before maturation, so that the endocrine cell clusters become scattered throughout the pancreatic tissue. 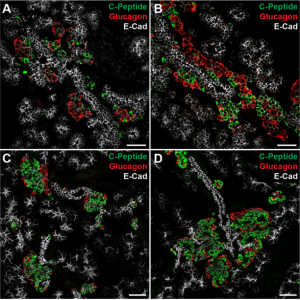 On p. 3744, Seung K. Kim and colleagues now uncover a mechanism controlling the migration of these cells and the subsequent positioning of islets. Using transgenic mouse lines and protein-soaked beads, they identify the semaphorin ligand Sema3a as a long-range guidance signal for migrating foetal islet cells. They show that endocrine cells express high levels of the Sema3a receptor neuropilin 2, allowing them to sense and transduce this chemoattractant. Intriguingly, this mechanism is similar to that which controls neuronal migration in the developing brain, and thus uncovers a conserved mechanism for directing the migration of progenitor cells during organogenesis.
On p. 3744, Seung K. Kim and colleagues now uncover a mechanism controlling the migration of these cells and the subsequent positioning of islets. Using transgenic mouse lines and protein-soaked beads, they identify the semaphorin ligand Sema3a as a long-range guidance signal for migrating foetal islet cells. They show that endocrine cells express high levels of the Sema3a receptor neuropilin 2, allowing them to sense and transduce this chemoattractant. Intriguingly, this mechanism is similar to that which controls neuronal migration in the developing brain, and thus uncovers a conserved mechanism for directing the migration of progenitor cells during organogenesis.
Decisions, decisions: cell fate choice in the early mammalian embryo
During the first week of mammalian development, the cells of the early embryo undertake two sequential cell fate decisions and segregate into the three lineages that make up the blastocyst. First, the trophectoderm (the precursor tissue to the placenta) becomes specified, occupying the outside of the embryo and surrounding uncommitted inner cell mass (ICM) cells. These subsequently segregate to Nanog-expressing epiblast (EPI; the embryo proper) and Gata6-expressing primitive endoderm (PE; yolk sac precursors).
In this issue, two research articles, one from the Ema lab and the second a collaboration between the Plusa and Piliszek labs, delve deeper into what governs the crucial EPI/PE cell fate specification event, which is well known to be reliant on FGF/ERK signalling. 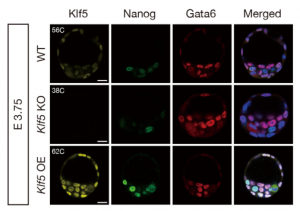 On p. 3706, Masatsugu Ema and colleagues provide evidence that the transcription factor Klf5 lies upstream of FGF/ERK signalling. They find that Fgf4 is upregulated in Klf5-knockout mouse embryos, which fail to specify EPI and can only generate PE. Conversely, when Klf5 is overexpressed, ICM cells fail to segregate, and continue to co-express both Nanog and Gata6. Furthermore, they find that Klf5 binds to the Fgf4 locus, suggesting that Fgf4 expression can be directly regulated by this transcription factor.
On p. 3706, Masatsugu Ema and colleagues provide evidence that the transcription factor Klf5 lies upstream of FGF/ERK signalling. They find that Fgf4 is upregulated in Klf5-knockout mouse embryos, which fail to specify EPI and can only generate PE. Conversely, when Klf5 is overexpressed, ICM cells fail to segregate, and continue to co-express both Nanog and Gata6. Furthermore, they find that Klf5 binds to the Fgf4 locus, suggesting that Fgf4 expression can be directly regulated by this transcription factor.
In the second study (p. 3719), Anna Piliszek et al. use a different model system – the rabbit – to investigate lineage specification in early embryos. 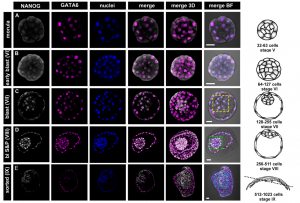 Unlike the mouse embryo, they detect co-expression of NANOG and GATA6 in late blastocysts, suggesting that mutual co-repression does not function in the initiation of lineage specification in rabbit. Furthermore, they show that inhibition of FGF signalling is not sufficient to expand the population of EPI cells, and although the population of PE cells is reduced, GATA6 expression is unaffected. These results indicate that although the key transcription factors are conserved in early mammalian embryogenesis, the way they function, and the way they interact with signalling pathways, may differ between species.
Unlike the mouse embryo, they detect co-expression of NANOG and GATA6 in late blastocysts, suggesting that mutual co-repression does not function in the initiation of lineage specification in rabbit. Furthermore, they show that inhibition of FGF signalling is not sufficient to expand the population of EPI cells, and although the population of PE cells is reduced, GATA6 expression is unaffected. These results indicate that although the key transcription factors are conserved in early mammalian embryogenesis, the way they function, and the way they interact with signalling pathways, may differ between species.
Taken together, these data add to our understanding of how the earliest cell fate decisions are taken in the mammalian embryo, and how they vary across evolution.
Plus…
Mitotic bookmarking in development and stem cells
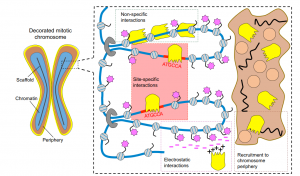
This Primer provides an overview of mitotic bookmarking processes in development and stem cells, highlighting how bookmarking factors can regulate cell identity and contribute to phenotypic flexibility and plasticity during development.
The three-dimensional genome
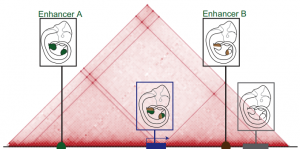
This Review summarizes the role of 3D chromatin architecture in organizing the regulatory genome and evaluates how its misfolding can lead to gene misexpression and disease.


 (No Ratings Yet)
(No Ratings Yet)



 (3 votes)
(3 votes)