![[Parameter-Settings] FileVersion= 2000 Date/Time = 0000:00:00 00:00:00 Date/Time + ms = 0000:00:00,00:00:00:000 User Name = TCS User Width = 1024 Length = 1024 Bits per Sample = 8 Used Bits per Sample = 8 Samples per Pixel = 3 ScanMode = xy Series Name = ptc_msl1p35_MMP1 p35 4d29.lei](https://thenode.biologists.com/wp-content/uploads/2016/02/DrosWing_LBarrio1-300x300.jpg)
Aneuploid cells—that is to say those with an abnormal number of chromosomes—are found in most human tumours.
A study conducted at IRB Barcelona on the fly Drosophila reveals how surviving aneuploid cells favour tumour development.
Barcelona, Thursday 9th February 2016.- A recent analysis of 43,205 human tumours unveiled that 68% of solid tumours are aneuploid, that is to say, they have an altered number of chromosomes. In recent years, scientists have attempted to clarify whether this aneuploidy contributes to tumour development or whether it is a co-lateral effect of the genomic instability of cancer cells, which increase the rate of mutations and the likelihood of cancer.
A study by the group headed by ICREA researcher Marco Milán, at the Institute for Research in Biomedicine (IRB Barcelona), published in this week’s issue of Developmental Cell provides details of the relationship between genomic instability, aneuploidy, and cancer.
The study, which has also involved the collaboration of ICREA researcher Angel R. Nebreda, in the Oncology’s programme at the same institute, explains how the molecular and cellular mechanisms triggered by aneuploid cells can give rise to tumours.
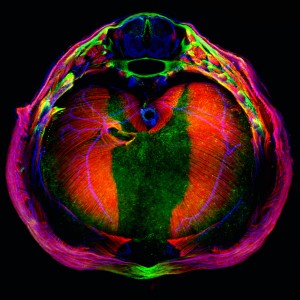
The research on aneuploidy and tumorigenesis has been performed using the wing primordia of the fruit fly Drosophila melanogaster as a model. This tissue is an epithelium organised into a single layer and that grows by 20 to 30,000 cells in a few days. Given these features, this tissue is an ideal system in which to generate genomic instability and to dissect the cell and molecular mechanisms that elicit aneuploid cells in a proliferating tissue.
Aneuploid cells: first step, suicide
The team of researchers observed that aneuploid cells first activate apoptosis (or programmed cell suicide). At the same time, in an attempt to counteract the imminent loss of cells, they send signals to neighbouring ones instructing them to divide and proliferate further to ensure the development of normal tissue—in this case the fly wing. Next, they also activate a series of DNA repair signals and also anti-tumour protection in order to prevent further aneuploidy.
“We have described the cascade of cell and molecular processes, and repair defence and compensation mechanisms which, simultaneously or sequentially, are triggered in and by aneuploid cells,” explains the postdoctoral researcher Marta Clemente, first author of the study.
But what happens if aneuploid cells manage to survive? After preventing the cells from dying, the researchers observed that the proliferation signals derived from aneuploid cells, which previously served to maintain healthy tissue, now favoured tumour development.
This study widens the Darwinian perspective of genomic stability in the development of cancer, “perhaps an incomplete view of the role of genomic stability in tumorigenesis” says Milán. Such a perspective is based on a random increase in tumour-promoting genes and a loss of tumour-supressing genes, which ultimately favours the tumour cell.
“Somehow the aneuploidy derived from this genomic instability also causes metabolic stress, which in turn leads to the expression of a series of signals that can enhance tumour growth and development”.
Given that aneuploidy is common to most cancers, Marco Milán considers that searching for treatments directed exclusively at removing aneuploid cells may provide a good strategy to tackle them.
“This basic biology research provides new information about the molecular links triggered by aneuploid cells, and this is the step prior to studying possible therapies to combat cancer,” says the IRB Barcelona researcher.
Reference article:
Gene dosage imbalance contributes to chromosomal instability-induced tumorigenesis
Marta Clemente-Ruiz, Juan M. Murillo-Maldonado, Najate Benhra, Lara Barrio, Lidia Pérez,
Gonzalo Quiroga, Angel R. Nebreda, and Marco Milán.
Developmental Cell (2016): doi: 10.1016/j.devcel.2016.01.008
This article was first published on the 9th of February 2016 in the news section of the IRB Barcelona website
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)

 (2 votes)
(2 votes)




![[Parameter-Settings] FileVersion= 2000 Date/Time = 0000:00:00 00:00:00 Date/Time + ms = 0000:00:00,00:00:00:000 User Name = TCS User Width = 1024 Length = 1024 Bits per Sample = 8 Used Bits per Sample = 8 Samples per Pixel = 3 ScanMode = xy Series Name = ptc_msl1p35_MMP1 p35 4d29.lei](https://thenode.biologists.com/wp-content/uploads/2016/02/DrosWing_LBarrio1-300x300.jpg)