The following is my response to the Nurse review on scientific funding call for evidence:
https://www.gov.uk/government/consultations/nurse-review-of-research-councils-call-for-evidence
Now he has lots of free time(!) having stepped down from running the Royal Society, Sir Paul remains the biggest name in scientific establishment, or at least the first name in the section marked ‘scientists’ in David Willett’s former permanent secretary’s phonebook (ah hem, I mean iPad). As such, he is conducting a massive review, following the previous massive review published last April:
https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/303327/bis-14-746-triennial-review-of-the-research-councils.pdf.
I don’t know (I am not RCUK; I can’t predict the future), but I bet the conclusion is along the lines of:
”This review notes that UK science has been excellent for most of its history. However, the obvious problem with the theory of evolution, the standard model of particle physics, all of medicine, and all of technology is that they did not have enough impact. Over the last decade, impact has been invented and now the world is better and more impactful. This has had no impact [ha!] on the quality of UK science, but has lead to the biggest scientific capital project in the history of Europe, that I was put in charge of. We need more impact, and analysis of impact. And impactfulness. Impact. Impact. Impact. The end. Impact.”
As such, EVERYBODY should respond to the call for comments. Stop what you are doing and respond. Now. Please. Once you have done that, if you can be bothered to or would actually like to read it, here is my response:
”Strategic decision-making
The ability of a government funding body, however well informed or advised by experts, to predict the future direction of scientific progress has proved demonstrably terrible throughout the entire history of science (arguably the history of government). As such, I have no objection to the current methods and practice of horizon scanning at RCUK (though my expertise very much only applies to the BBSRC and the MRC); they are probably better than they have ever been. Rather, I object enormously to the importance they are bestowed with. Decision-making in relation to scientific strategy is something on which research funders, with the exception of those with a very specific scientific remit (eg. small medical charities dedicated to particular conditions), should place very little importance. Rather, scientific direction should be the organic result of scientific progress, which is completely unpredictable. Funders should not pretend they can predict it. They cannot.
The practice of systematically concentrating research in strategic areas has developed hand-in-hand with an emphasis on economic and other less pernicious forms of ‘impact’ – ‘‘describe how your research will contribute to the economic performance of UK PLC’’. This is often justified on the basis that taxpayers money should be accounted for with democratic imperative. This is a very noble goal, but one which is entirely misplaced in the context of science. It has developed extensively over the last decade, largely at the imperative of ministers and senior civil servants (according to the triennial review of research councils – section 116) in a manner that I would argue counteract the Haldane Principle. No voter, even amongst scientists, makes political decisions on the basis of scientific funding. This is quite as it should be. Science is an activity that by its very nature is entirely undemocratic and entirely meritocratic. Scientific discoveries that change the world do so irrespective of whether they were popular when made.
In contrast, the emphasis on impact makes the process of funding in the UK far more opaque, since no one (not even the research councils) can judge it sensibly, as acknowledged in the triennial review – sections 118-124. (Unfortunately, the civil servants authoring the review suggest in sections 125 and 126 that because it is not judged sensibly at present, such measurement should receive more emphasis. Predictably, they do not suggest how). Most importantly, the emphasis on impact also means that research proposals are not judged based purely upon quality. This is ludicrous and profoundly damaging to the future of science in the UK. It is quite rightly ridiculed by American and European colleagues.
Balance of funding portfolio
The high proportion of the total funding pot available through the BBSRC and MRC for council-specified ‘strategic target areas’ has, over the period of my scientific career (2005 to date) got progressively larger in absolute and relative terms. This has had the direct result of concentrating research in a small number of areas, and in a small number of laboratories, that are deemed of high importance by a small number of (albeit well qualified) people. This is a model of science funding that would systematically overlook the enormous curiosity-driven, and entirely unpredictable, world-changing findings that characterise the world class UK science base since the end of the second world war, and indeed all great science in every place and every time.
Secondly, the modern practice of making even the dwindling curiosity-driven responsive mode part of the portfolio subject to judgement against the very same strategic objectives makes the system not only enormously opaque, but means that as it currently operates, the system is profoundly unmeritocratic. This is ludicrous, as anyone with even a partial grasp of the scientific process and its history ought to realise. As currently constituted, the entire funding scheme is based upon a profoundly mistaken philosophical premise: that the scientific process is predictable. It is not. Even in modern times we have optogenetics because people were studying unfashionable algal ion channels; we have CRISPR because people were studying odd ‘non-functional’ regions of strange bacterial and archael genomes. It is not possible, even for the finest minds, to predict the future direction of science. HM government certainly should not have an entire system predicated upon doing so itself.
Effective ways of working
The current career structure in science is absolutely appalling, and has got steadily worse over the course of my career (2005 to date). In contrast to scientific strategy, it is precisely the sort of thing that should be the subject of strategic oversight by governments. In a sad irony, while scientific direction has been the subject of increasing beaurocarcy, the career structure has been left to grow organically with no planning. This has resulted in the overproduction of people with almost no employment rights and even worse career prospects in science precisely at the age at which they should be able to start families. It is why the brightest graduates are no longer choosing science as a career, and why scientists are advising them against doing so. Sadly predictably, it underlies the appalling lack of sexual and ethnic diversity amongst scientific workforces, even at junior levels: we are almost at the point where it is necessary to have independent wealth to pursue a scientific career. Women, quite correctly, judge in droves that it is not a career for them since it systematically discriminates against people who are not heavily engaged in self-promotion and able to commit a very long working week. This means that increasing numbers of scientists who do achieve permanent positions do so at the expense of having a family, leading to even less recognition for family commitments in the workforce: a vicious circle that benefits no-one.
There is a good argument that the scientific pyramid is responsible for providing highly qualified people for the knowledge economy of the future. I wholeheartedly agree. However, the current system that prevents the brightest talent from progressing in their career in their late 20s and 30s (and increasingly 40s) ie. the age at which they would be parents, negates this. PhDs are excellent qualifications that are an inherent good for society and the economy. A sensible system would encourage education to PhD level. However, the postdoc stage is not a ‘training’ period, and should not be treated as such. There should be far fewer postdocs, with each being granted longer contracts and far more security. To be clear, I am not advocating less postdoc hours in the work force. Just less postdocs. Each would have much more time. In this way, science would be the desireable career it ought to be. Just as importantly, it would also, of course, drastically increase the quality of the science being done. Pressure does not lead to better science; curiosity does.”
 (10 votes)
(10 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (2 votes)
(2 votes)
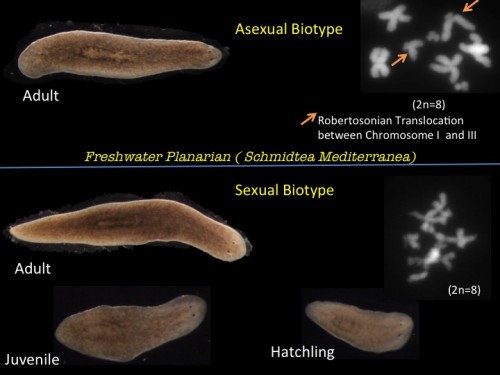
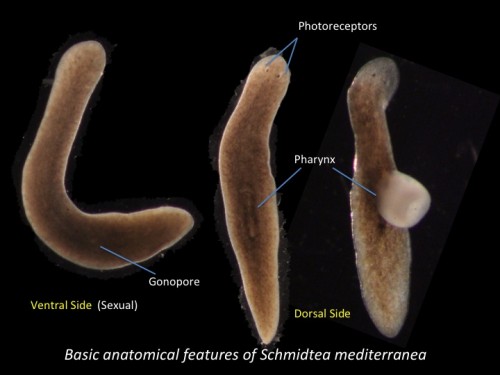
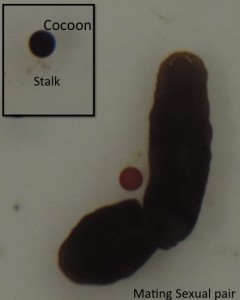
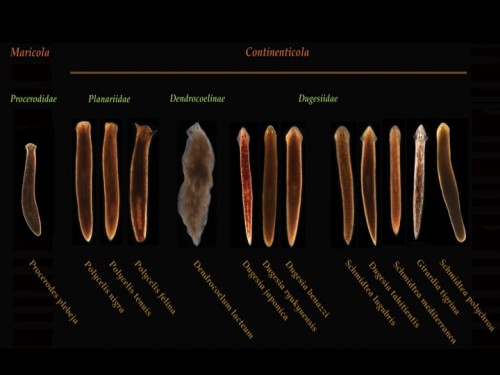

 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series  (16 votes)
(16 votes)