The MRC National Institute for Medical Research has celebrated its centenary in 2014. On 1st April 2015 NIMR ceased to exist, as it became part of the new Francis Crick Institute. To commemorate this event, I proposed to produce a textile artwork as a lasting memento of NIMR. Each current research lab, support section and club was invited to create a piece of artwork on a small square of cloth. They were then combined into a single work that celebrates the science, life and ethos of NIMR.

Image 1 – a full view of the completed NIMR canvas, which consists of one hundred individual pieces. (click to see bigger image)
Below, I present the patches created by the developmental biologists from NIMR. These patches are beautifully crafted with an impressive array of imagination and creativity.
NIMR is home to many world renowned developmental biologists, past and present, among them is Rosa Beddington. Rosa was an influential figure and a gifted artist. One of her drawings inspired two pieces of artwork for the canvas.
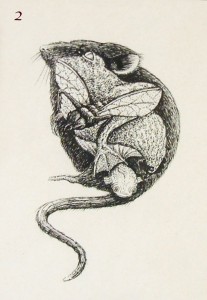
Image 2 – drawing by Rosa Beddington to illustrate the conservation of developmental pathways between the mouse, frog, fruit fly and fish.

Image 3 – hand stitched by Alan Palmer from Biological Services. This is an adaption of the Rosa Beddington sketch and represents the diversity of animal models used at NIMR and the adaptability of Biological Services in providing the best possible care and welfare for all animals.

Image 4 – mixed media artwork by Patti Biggs at the Library. Interestingly, Patti added her own interpretation of Rosa’s drawing by turning the tail of the mouse into a nematode worm, another well known model organism in developmental biology.
Below are some pieces created by current NIMR labs in the field of developmental biology. These artworks were produced with various techniques including fine needlework such as cross stitches, applique, beading as well as painting and printing.

Image 5 – Jim Smith lab by Elsie Place, Alex Eve, Melissa Estima and Rita Monteiro, depicting the SuperFrog. ‘SuperFrog’ was inspired by past and current research in Smith lab as an energetic action hero. The boxed ‘T’ symbol on SuperFrog’s chest represents Brachyury and other members of the T-box transcription factor family.

Image 6 – Robin Lovell-Badge lab by Rosalyn Flower. The lab studies how the vertebrate gonad develops to become a testis in males or an ovary in females, a process known as sex determination. This is depicted on the canvas as a battle between the sexes. On the left is a male mouse and a cartoon of the testis with its characteristic cords and on the right is a female mouse and a cartoon of the ovary with its characteristic follicles.

Image 7 – Robin Lovell-Badge lab by Robin Lovell-Badge and Karine Rizzoti. The lab found Sox2 and Sox9 have pleiotropic roles during mouse development and in stem cells in the adult. For example, SOX2 is essential for pluripotent cells in the early embryo, such as those of the early epiblast and inner cell mass of the blastocyst (top left), embryonic and other pluripotent stem cells in culture (bottom left), progenitors in the developing hypothalamo-pituitary axis (top right), and neural stem cells (NSCs) in vitro and in vivo, including in specialised niches in the adult brain (bottom right). In the embryo, maintenance of early pituitary progenitors (top left, blue) depends on signals from the overlying ventral diencephalon (green). In the adult brain NSC niche (bottom right), in the lateral ventricles, ependymal cells (green), and the microvasculature (red) contribute signals that help to regulate activation, proliferation and differentiation of the NSCs.
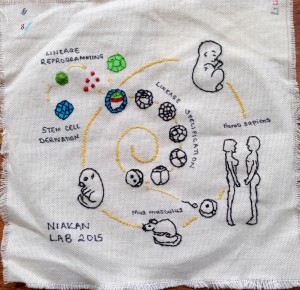
Image 8 – Kathy Niakan lab by Sissy Wamaitha. The patch illustrates the foundations for the models (mouse and human) used to understand mammalian development in the lab. Elements of the embryo development timeline from a standard figure in the lab show the series of cell divisions culminating in the formation of the blastocyst in early mouse and human development; stem cells and later stages of development (foetus and adult) are also included.
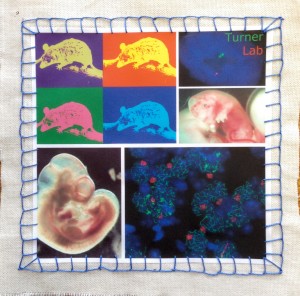
Image 9 – James Turner lab by Shantha Mahadevaiah, Daniel Snell and Fanny Decarpentrie. The piece encapsulates their work on sex chromosomes and development in mammals using the model organisms Mus musculus, the house mouse, and Monodelphis domestica, the short-tailed grey opossum. From top left, clockwise: opossum, in pop-art style; RNA FISH using opossum embryonic fibroblast; E13 opossum embryo; chromosomal spread from mouse testis; and mouse embryo at E10.5.

Image 10 – James Briscoe lab by Mina Gouti. The canvas was inspired from her recent work showing the generation of neuromesodermal progenitors in the culture dish from mouse and human embryonic stem cells following the cues from mouse embryonic development. Cells of different colours in a petri dish represent in vitro generated neuromesodermal progenitors. These are the building blocks of the spinal cord and most of the muscle tissue in our body. The different colours show the potential of these cells to form both the different spinal cord progenitor domains as well as the adjacent somites.

Image 11 – David Wilkinson lab by Angela Cheung, Megan Addison, Hannah Stanforth, Harriet Taylor, David Wilkinson, Zhonglin Wu and Qiling Xu. It illustrates the focus of the lab in the mechanisms and roles of cell segregation and boundary formation in the developing hindbrain. The design depicts various themes of the lab’s work. At the top are zebrafish embryos at four different stages of development. In the middle, the segmented organization of the hindbrain is shown, with segments in green and red, boundary cells in yellow, and fgf20-expressing neurons in gold. Below are green- and red-labelled HEK293 cells expressing Eph receptor and ephrin that are used to investigate mechanisms of cell segregation.

Image 12 – Rainbow fish by Qiling Xu, depicting a transgenic rainbow zebrafish, red eye selection and NIMR workshop-made moulds used for live imaging. It shows appreciation for the animal facility, biological service and aquatic staff at NIMR. So long, “thanks for all the fish!” A quote from Douglas Adams.

Image 13 – Greg Elgar & Mike Gilchrist labs by Lilly Hunt, Greg Elgar, Mike Gilchrist, Johanna Fischer, Htoo Wai, Stefan Pauls, Laura Doglio, Boris Noyvert, Joe Grice, Brook Cooper, Ian Grant, Nick Owens, Ilya Patrushev, Elena DeDomenico and Rosa Gomes Faria. This is a joint patch from the two labs as they work closely together, sharing lab space and ideas. The Gilchrist lab works with Xenopus embryos whereas the Elgar lab uses zebrafish, and both model organisms are incorporated onto the canvas. The Warhol style in his famous Marilyn Monroe images was used to create a striking image.

Image 14 – Tim Mohun lab by Christina McGuire. The foundation of the piece is a 3 micron thick HREM (high resolution episcopic microscopy) image of a 14.5 day old mouse embryo which has been coloured and printed onto the canvas. The embryo’s developing structures have been highlighted using metallic paint, silver thread, beads, glitter and angelica fibres, accentuating the beauty of the embryo. The image was chosen as it represents both the Mohun group research on heart development, and the Wellcome Trust funded DMDD (Deciphering the Mechanisms of Developmental Disorders) programme which aims to study “embryonic lethal” mouse mutations, and begin to understand why their mutation has such profound effects on embryo development and survival.

Image 15 – Vassilis Pachnis lab by Sarah McCallum. The canvas represents a cross section of the gut showing the Enteric Nervous System (ENS) in blue with the myenteric and submucosal plexus in the gut wall, and the neuronal fibres entering the villi.

Image 16 – Siew-Lan Ang lab by Siew-Lan Ang and Shabana Khan, depicting midbrain dopaminergic neurons and their amazing journey to the forebrain.

Image 17 – Francois Guillemot lab by Debbie Van den Berg, Noelia Urban, Isabelle Blomfield and Koji Oishi, showing the adult neural stem cells, with their typical long radial processes, in various states of cell activation. They are depicted either in their natural niche, the dentate gyrus of the hippocampus, where they contribute to memory formation, or sprouting from a tissue culture dish, where their response to various stimuli can be studied in a detailed and controlled manner.

Image 18 – Sila Ultanir lab by Suzanne Claxton and Lucas Baltussen, showing a hard working minion. The kinase reaction that replaces the eye stands for the lab’s focus on kinases in a neurodevelopmental background depicted by the surrounding neurons.
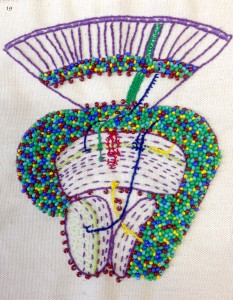
Image 19 – Iris Salecker lab by Iris Salecker, Holger Apitz, Kathleen Dolan, Richard Kaschula, Benjamin Richier and Nana Shimosako. This patch shows an adult visual system of the fruit fly – a model to understand how neural circuits assemble during development. It contains clues to the past and ongoing research. Stippled lines reveal the organization of axonal and dendritic neurites into synaptic layers. Beads in “Flybow” colors represent the cell bodies of neurons and glia. We included our favorite cell types: photoreceptor axons R1-R8 (green), lamina neuron L3 (blue, left), Nana’s medulla neuron Tm20 (blue, right), Richard’s amacrine-like medulla neuron Dm3 (yellow), Benjamin’s and Kathleen’s astrocyte-like medulla neuropil glia (red, center) and Holger’s lobula plate neuron T4 (yellow). Light green symbolizes the expression of one of our molecules of interest, the chemoattractant guidance cue Netrin-B.

Image 20 – Jean-Paul Vincent lab by Zoe Vincent and Iris Salecker, showing a cubist version of Drosophila, used to investigate signaling between cells and to determine how signalling contributes to growth, patterning and cell death.
Further information on the history of NIMR and the Developmental Biology at NIMR can be found in the following links:
http://www.historyofnimr.org.uk/100-years-of-science-for-health-the-book/
http://www.historyofnimr.org.uk/files/2014/07/chapter21.pdf
 (8 votes)
(8 votes)
 Loading...
Loading...


 (1 votes)
(1 votes)





 (No Ratings Yet)
(No Ratings Yet)



















 (8 votes)
(8 votes)