Heather Hendrickson (Senior Lecturer in Molecular Bioscience, Massey University)
I have had my personal ups and downs with live performance. I was 4 years old when, convinced I was a dance prodigy, my enthusiastic whirling became frenetic and I flung myself directly at my grandmother, landing in a humiliated heap at her feet. Despite the signs, it still took me years to recognize that dance performance was not to be my destiny. I was fortunate enough to find science. Today I am a Senior Lecturer in Molecular Bioscience at Massey University in New Zealand. The research in my laboratory focuses on the evolution of bacteria, DNA replication and segregation and bacteriophage discovery. For the past two years I have also been the Chair of the Outreach Committee for my Institute. This position put me in touch with our campus External Relations team, the group that designs and implements strategies for university engagement with the public.
When a national TV station in our city came looking for a science news correspondent, our external relations team asked me if I was interested in trying this. As a result, I have made monthly TV appearances on a nationwide morning program in New Zealand for the past two years. We are very lucky to have this forum for a weekly science update, with five academics from around the country talking to the public about science for 5 minutes each week, in a live interview format . This has been a huge learning opportunity for me and has opened up a wealth of other opportunities in science communication, including radio interviews, a podcast, public talks, other TV programs and filming for a documentary.
 In my natural environment, the bench.
In my natural environment, the bench.
What is it like to do science communication on Live TV?
The morning program, First Line, has a large share of the pre-work family viewing audience: young families, mature couples and singles who want a dose of news before heading to school or work. The goal of the station is therefore to provide viewing material that will keep them tuned in.
As with any science outreach, the audience comes first, so I choose my topics carefully. Perfect stories are those about recent science topics that both I and that the public find interesting. The story should have an “aha!” factor. If the stories are good the rest is easy, as long as you have a store of natural enthusiasm for the topics. I read widely, check online sources like Science Daily Headlines, Twitter and even Facebook for recent trending science news. I am also subscribed as a journalist with the Science Media Center in New Zealand. This is an invaluable resource, as I receive updates about embargoed press releases that I can research in order to present the most “up to date” science news available.
Once I have chosen 3-5 stories, I send abstracts, articles on-line, videos and anything else that might be helpful to the producer, the day before we go on air. The producer and I choose three stories from my prescreened ideas. Selected stories have been everything from bacteriophage therapy and antibiotic resistance (close to my heart) to advances in robotics and climate change (very far afield). When in doubt I call on old friends or other experts to put things in context for me.
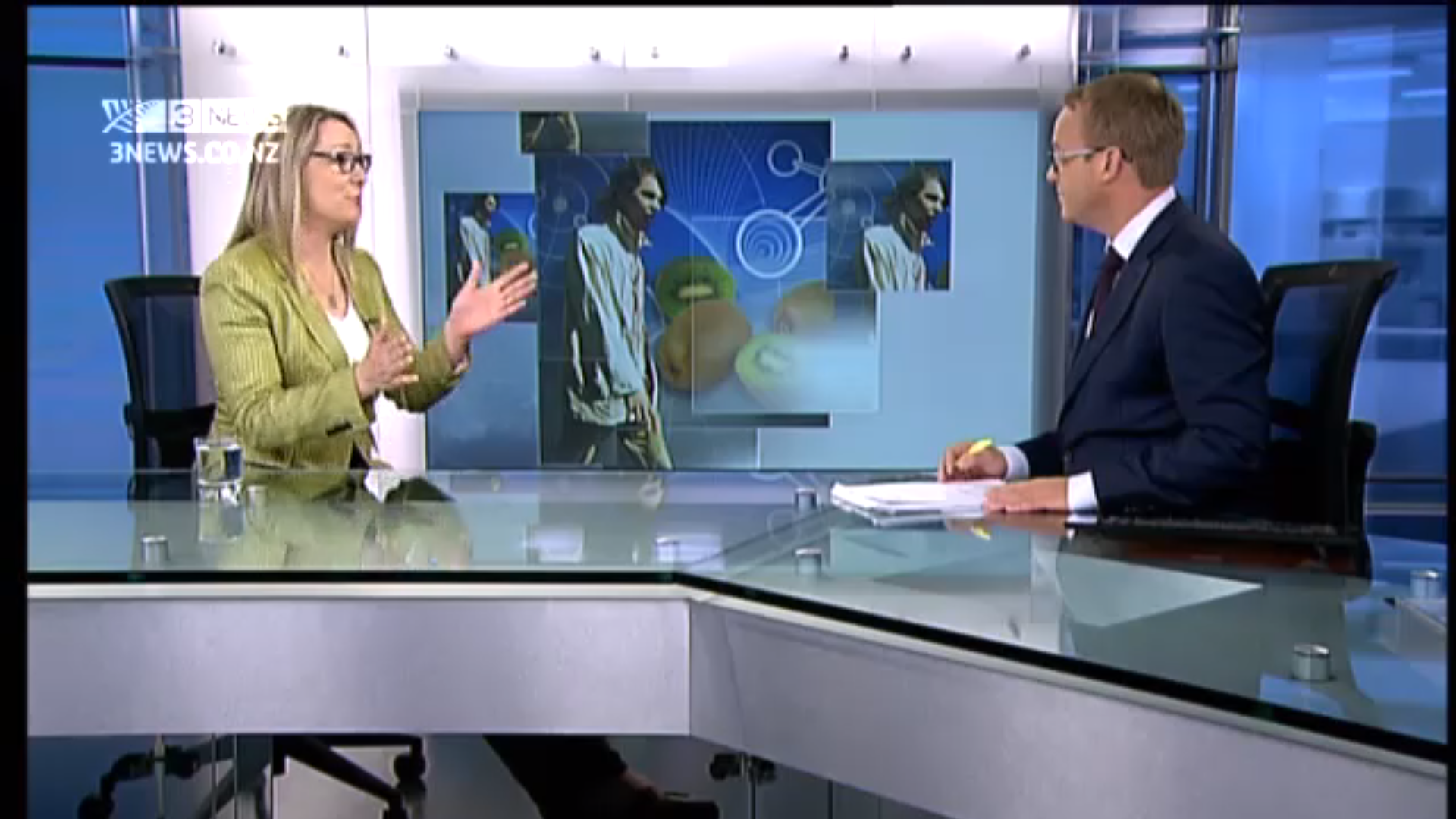
On First Line, TV3, New Zealand. Note the video monitor behind us has both kiwifruits and a zombie!
Once the topics are selected the task is to choose my big concepts and simplify, simplify, simplify. I have about 1.5 minutes of discussion for each topic, and I must assume my audience has no knowledge of acronyms, techniques, or context. This changes what I choose to talk about. If the topic is relevant then the language should come fairly naturally. Complex ideas must be translated into normal language.
By the morning of the interview, I have in mind a summary of what the work is, why it is important and what comes next for this field. I have very rarely been given questions in advance, so I have to be ready to guide a strange or off topic question to a productive point.
I arrive at the studio 45 minutes before go on air, and head into make up for a 15 minute hair and make up session with the amazing professionals on staff. About 5 minutes before my segment I am invited on set, say hello to the hosts and get my microphone subtley installed.
Top Tips for going live:
1) Don’t wear things that make noise, the mics are sensitive!
2) Sit up straight but lean slightly forward to physically engage the interviewer.
3) Avoid the temptation to look at the tele-prompters or the camera.
4) Smile and maintain eye contact with the interviewer, even at the end of the interview.
5) If a question seems strange, simply ask for clarification. This is a conversation!
6) Keep things simple, engaging and light-hearted (where appropriate).
7) Obstinate hosts can drive ratings so don’t take things personally. You are there for the audience (not the interviewer) so play along, but know your limits in advance.
 Me and Professor Steve Pointing doing the end of year wrap up, December 2014.
Me and Professor Steve Pointing doing the end of year wrap up, December 2014.
How is Live TV science outreach different?
The biggest difference between TV-based outreach and classroom-based outreach is that when you do face to face outreach with students or the public you can see the effects immediately. Sometimes their faces light up with recognition or understanding, and other times you can hear expressions of awe. I think that for many of us this is one of the most rewarding parts of science communication. This does not happen when you do science communication on live TV. When I walk out of the TV station after my five minutes on air, I don’t know who might have seen the spot or what they thought. I have occasionally received appreciative e-mails from viewers afterwards, but these are few and far between .
Should you get involved in live TV science outreach?
The public funds our work, so we owe it to them to tell them about discoveries and progress that are relevant to their lives. I also remind myself that there are little science nerds out there that don’t know it yet. Maybe some of them even think that they are destined to be great dancers! I like the idea of inspiring the kids of today to be the scientist of the future.
So if you think that live TV outreach sounds fun, you should go for it. Not sure if you can pull it off? Do a trial run with someone from your external relations or press team on campus. These groups will have direct experience with live television, live radio or print media, and they will have great tips and leads for you to make contact with journalists. They are also likely to know the local media personalities and can give you an idea of what to expect.
There are also great training sessions and resources online: http://sciencemediasavvy.org/
For a video of me on TV covering bacteriophages and human induced climate change follow this link: http://www.3news.co.nz/environmentsci/using-viruses-to-kill-harmful-bacteria-2013052909#axzz3UQ33Rp4a This was one of the first interviews I did and at one point, the interviewer asks if climate change wasn’t “getting better”, which surprised me.
You can find more links to my appearances on First Line in my Science Communication page: http://microbialevolution.massey.ac.nz/HHmedia.html
 This post is part of a series on science outreach. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on science outreach. You can read the introduction to the series here and read other posts in this series here.
 (4 votes)
(4 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (4 votes)
(4 votes) (6 votes)
(6 votes) The medium-sized spiny neurons, the main projection neurons of the striatum, are generated in the lateral ganglionic eminence (LGE) and degenerate in the early stages of Huntington’s disease (HD) – for which no pharmacological treatment is yet available. Hence, an efficient way to derive striatal neurons is crucial for disease modelling, drug development and cell-replacement therapy. Striatal neurons have previously been generated from human pluripotent stem cell (hPSC)-derived neural progenitors treated with sonic hedgehog (SHH), or SHH plus Wnt pathway inhibition. Now, Meng Li and co-workers (p.
The medium-sized spiny neurons, the main projection neurons of the striatum, are generated in the lateral ganglionic eminence (LGE) and degenerate in the early stages of Huntington’s disease (HD) – for which no pharmacological treatment is yet available. Hence, an efficient way to derive striatal neurons is crucial for disease modelling, drug development and cell-replacement therapy. Striatal neurons have previously been generated from human pluripotent stem cell (hPSC)-derived neural progenitors treated with sonic hedgehog (SHH), or SHH plus Wnt pathway inhibition. Now, Meng Li and co-workers (p.  Neural tube closure occurs through highly orchestrated cell shape changes mediated by actin dynamics. Its failure results in some of the most common and severe human congenital malformations. Cofilin 1, an actin-depolymerising protein, is known to be involved in neural tube closure but its precise functions had not been elucidated. In this study (p.
Neural tube closure occurs through highly orchestrated cell shape changes mediated by actin dynamics. Its failure results in some of the most common and severe human congenital malformations. Cofilin 1, an actin-depolymerising protein, is known to be involved in neural tube closure but its precise functions had not been elucidated. In this study (p.  On p.
On p.  In the second study (p.
In the second study (p.  The two most influential ideas in the field of pattern formation are those of Alan Turing’s ‘reaction-diffusion’ and Lewis Wolpert’s ‘positional information’. Much has been written about these two concepts but some confusion still remains, in particular about the relationship between them. Here, Jeremy Green and James Sharpe address this relationship and propose a scheme of three distinct ways in which these two ideas work together to shape biological form. See their Hypothesis article on p.
The two most influential ideas in the field of pattern formation are those of Alan Turing’s ‘reaction-diffusion’ and Lewis Wolpert’s ‘positional information’. Much has been written about these two concepts but some confusion still remains, in particular about the relationship between them. Here, Jeremy Green and James Sharpe address this relationship and propose a scheme of three distinct ways in which these two ideas work together to shape biological form. See their Hypothesis article on p. 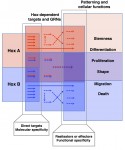 Hox genes encode homeodomain transcription factors that control morphogenesis and have established functions in development and evolution.Here, Yacine Graba and colleagues discuss the molecular and cellular mechanisms underlying the diverse and context-dependent functions of Hox transcription factors during morphogenesis and organogenesis. See the Review article on p.
Hox genes encode homeodomain transcription factors that control morphogenesis and have established functions in development and evolution.Here, Yacine Graba and colleagues discuss the molecular and cellular mechanisms underlying the diverse and context-dependent functions of Hox transcription factors during morphogenesis and organogenesis. See the Review article on p. 



