Here are the research highlights from the current issue of Development:
Directed differentiation: Nodal steps forward
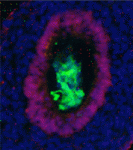 The directed differentiation of pluripotent stem cells into endodermal derivatives, including insulin-producing pancreatic β cells, has considerable clinical promise in cell replacement therapies. The first step in this process is the conversion of pluripotent stem cells into definitive endoderm (DE). Here (p. 675), Douglas Melton and colleagues investigate the endodermal populations generated from mouse embryonic stem cells treated with Nodal (which is required for in vivo development of DE) or Activin A (which is thought to mimic Nodal activity). These TGFβ family members use the same signalling pathways but, although the researchers show that Nodal- and Activin-derived DE cells have similar gene expression patterns, Nodal-derived endoderm contributes much more efficiently to embryonic endoderm upon transplantation into the gut endoderm of mouse embryos. Importantly, this functional difference between Nodal- and Activin-derived endoderm extends to the subsequent development of pancreatic progenitors in vitro and maturation into insulin/c-peptide-expressing cells in vivo. These data provide a firm basis for the derivation of insulin-producing cells for disease modelling and cell therapy.
The directed differentiation of pluripotent stem cells into endodermal derivatives, including insulin-producing pancreatic β cells, has considerable clinical promise in cell replacement therapies. The first step in this process is the conversion of pluripotent stem cells into definitive endoderm (DE). Here (p. 675), Douglas Melton and colleagues investigate the endodermal populations generated from mouse embryonic stem cells treated with Nodal (which is required for in vivo development of DE) or Activin A (which is thought to mimic Nodal activity). These TGFβ family members use the same signalling pathways but, although the researchers show that Nodal- and Activin-derived DE cells have similar gene expression patterns, Nodal-derived endoderm contributes much more efficiently to embryonic endoderm upon transplantation into the gut endoderm of mouse embryos. Importantly, this functional difference between Nodal- and Activin-derived endoderm extends to the subsequent development of pancreatic progenitors in vitro and maturation into insulin/c-peptide-expressing cells in vivo. These data provide a firm basis for the derivation of insulin-producing cells for disease modelling and cell therapy.
Unpaired Sox17 models biliary atresia
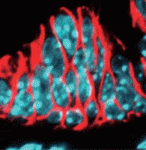 Congenital biliary atresia is an incurable disease of newborn infants that is characterised by deformation of the gallbladder and biliary duct system. Yoshiakira Kanai and co-workers now report (p. 639) that haploinsufficiency of Sox17 in C57BL/6 background mice provides a genetic model for this poorly understood condition. The researchers show that SOX17, a transcription factor that is required for definitive endoderm development in various vertebrate species, is expressed at the distal edge of the gallbladder primordium during gallbladder and bile duct development. In Sox17+/− C57BL/6 embryos, cell-autonomous defects in the proliferation and maintenance of the gallbladder/bile duct epithelia lead to epithelial cell detachment from the luminal wall, bile duct atresia (blockage), bile leakage and inflammation in the bile ducts and liver at late foetal stages. These results suggest that SOX17 has a dose-dependent function in the morphogenesis and maturation of gallbladder and bile duct epithelia during late organogenesis and provide new insights into the pathogenesis of congenital biliary atresia.
Congenital biliary atresia is an incurable disease of newborn infants that is characterised by deformation of the gallbladder and biliary duct system. Yoshiakira Kanai and co-workers now report (p. 639) that haploinsufficiency of Sox17 in C57BL/6 background mice provides a genetic model for this poorly understood condition. The researchers show that SOX17, a transcription factor that is required for definitive endoderm development in various vertebrate species, is expressed at the distal edge of the gallbladder primordium during gallbladder and bile duct development. In Sox17+/− C57BL/6 embryos, cell-autonomous defects in the proliferation and maintenance of the gallbladder/bile duct epithelia lead to epithelial cell detachment from the luminal wall, bile duct atresia (blockage), bile leakage and inflammation in the bile ducts and liver at late foetal stages. These results suggest that SOX17 has a dose-dependent function in the morphogenesis and maturation of gallbladder and bile duct epithelia during late organogenesis and provide new insights into the pathogenesis of congenital biliary atresia.
Embryonic DNA methylation without Dnmt3L
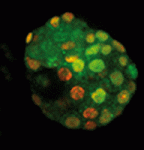 During embryogenesis and gametogenesis, the DNA methyltransferases Dnmt3A and Dnmt3B establish the genome-wide methylation patterns that are essential for mammalian development and reproduction. The catalytically inert Dnmt3-like (Dnmt3L) is known to regulate de novo methylation in the germline but does it function in the early embryo? Déborah Bourc’his and colleagues have been investigating this question and, on p. 562, they report that, although mouse embryos initially contain a maternal store of Dnmt3L, the protein is rapidly degraded. The researchers show that zygotic Dnmt3L deficiency slows down the rate of de novo methylation in the embryo by affecting methylation density at some, but not all, genomic sequences. Importantly, however, Dnmt3L is not strictly required for de novo methylation in the embryo because methylation patterns are eventually established in its absence, possibly through upregulation of Dnmt3A. De novo methylation can therefore be achieved in vivo without Dnmt3L, which suggests that early mouse embryos are more plastic than the germline in terms of how they acquire de novo methylation patterns.
During embryogenesis and gametogenesis, the DNA methyltransferases Dnmt3A and Dnmt3B establish the genome-wide methylation patterns that are essential for mammalian development and reproduction. The catalytically inert Dnmt3-like (Dnmt3L) is known to regulate de novo methylation in the germline but does it function in the early embryo? Déborah Bourc’his and colleagues have been investigating this question and, on p. 562, they report that, although mouse embryos initially contain a maternal store of Dnmt3L, the protein is rapidly degraded. The researchers show that zygotic Dnmt3L deficiency slows down the rate of de novo methylation in the embryo by affecting methylation density at some, but not all, genomic sequences. Importantly, however, Dnmt3L is not strictly required for de novo methylation in the embryo because methylation patterns are eventually established in its absence, possibly through upregulation of Dnmt3A. De novo methylation can therefore be achieved in vivo without Dnmt3L, which suggests that early mouse embryos are more plastic than the germline in terms of how they acquire de novo methylation patterns.
Mending a broken heart
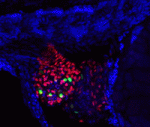 Human hearts do not regenerate after a heart attack because adult mammalian cardiomyocytes proliferate poorly in response to injury. By contrast, zebrafish regenerate heart muscle after trauma by inducing cardiomyocyte proliferation. Studies of zebrafish heart regeneration might, therefore, identify ways to repair damaged human hearts. Here (p. 660), Wen-Yee Choi and co-workers develop a surrogate model for zebrafish heart regeneration that uses fluorescent ubiquitylation-based cell cycle indicator (FUCCI) technology to visualise cardiomyocyte proliferation in live zebrafish embryos. The researchers generate transgenic lines in which heart-specific promoters drive the expression of G1 and S/G2/M FUCCI probes and use these lines to identify several small molecules that alter cardiomyocyte proliferation during heart development. These molecules act via the Hedgehog, IGF or TGFβ signalling pathways, they report. Moreover, the researchers show, the same pathways are activated in regenerating zebrafish cardiomyocytes, and their pharmacological manipulation alters cardiomyocyte proliferation during adult heart regeneration. Future use of this new screening system may identify molecules with the potential to improve human heart regeneration.
Human hearts do not regenerate after a heart attack because adult mammalian cardiomyocytes proliferate poorly in response to injury. By contrast, zebrafish regenerate heart muscle after trauma by inducing cardiomyocyte proliferation. Studies of zebrafish heart regeneration might, therefore, identify ways to repair damaged human hearts. Here (p. 660), Wen-Yee Choi and co-workers develop a surrogate model for zebrafish heart regeneration that uses fluorescent ubiquitylation-based cell cycle indicator (FUCCI) technology to visualise cardiomyocyte proliferation in live zebrafish embryos. The researchers generate transgenic lines in which heart-specific promoters drive the expression of G1 and S/G2/M FUCCI probes and use these lines to identify several small molecules that alter cardiomyocyte proliferation during heart development. These molecules act via the Hedgehog, IGF or TGFβ signalling pathways, they report. Moreover, the researchers show, the same pathways are activated in regenerating zebrafish cardiomyocytes, and their pharmacological manipulation alters cardiomyocyte proliferation during adult heart regeneration. Future use of this new screening system may identify molecules with the potential to improve human heart regeneration.
Eyeing up proneural bHLH factors
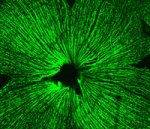 During retinal development, seven retinal cell types are specified from a common pool of retina progenitor cells (RPCs). Several proneural basic helix-loop-helix (bHLH) transcriptional regulators, including Atoh7 and Neurod1, direct the intrinsic programming of RPCs but how do individual bHLH factors influence RPC fate? On p. 541, William Klein and colleagues ask whether replacing one bHLH gene with another redirects the fate of RPCs. Previously, the researchers showed that Neurod1 can replace the function of Atoh7 in specifying retinal ganglion cells (RGCs), which suggests that Atoh7-expressing RPCs are pre-programmed to produce RGCs. Now, they report that insertion of Atoh7 into the Neurod1 locus reprogrammes Neurod1-expressing RPCs, which normally produce amacrine and photoreceptor cells, into RGCs. Thus, Atoh7 acts dominantly to specify an RGC fate. The researchers also identify an Atoh7-dependent enhancer within its target gene Nrxn3 that is used by Atoh7 but not by Neurod1 in the developing retina. Together, these results provide new insights into the specification of retinal cells by proneural bHLH factors.
During retinal development, seven retinal cell types are specified from a common pool of retina progenitor cells (RPCs). Several proneural basic helix-loop-helix (bHLH) transcriptional regulators, including Atoh7 and Neurod1, direct the intrinsic programming of RPCs but how do individual bHLH factors influence RPC fate? On p. 541, William Klein and colleagues ask whether replacing one bHLH gene with another redirects the fate of RPCs. Previously, the researchers showed that Neurod1 can replace the function of Atoh7 in specifying retinal ganglion cells (RGCs), which suggests that Atoh7-expressing RPCs are pre-programmed to produce RGCs. Now, they report that insertion of Atoh7 into the Neurod1 locus reprogrammes Neurod1-expressing RPCs, which normally produce amacrine and photoreceptor cells, into RGCs. Thus, Atoh7 acts dominantly to specify an RGC fate. The researchers also identify an Atoh7-dependent enhancer within its target gene Nrxn3 that is used by Atoh7 but not by Neurod1 in the developing retina. Together, these results provide new insights into the specification of retinal cells by proneural bHLH factors.
Body elongation goes with the flow
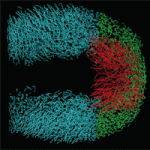 The tailbud is the posterior leading edge of the growing vertebrate embryo. Now, by measuring the three-dimensional cell flow field of the zebrafish tailbud, Scott Holley and co-workers reveal a posterior flow within the tailbud that reflects ordered collective cell migration (p. 573). They identify a transition in tissue fluidity at the tailbud tip where there is a decrease in the coherence of the cell flow but no alterations of cell velocities. Inhibition of Wnt or Fgf signalling reduces the coherence of the flow, but affects trunk and tail extension differently. By using computer simulations to interpret these complex phenotypes, the researchers show that a decrease in the coherence of the flow combined with a normal flow rate leads to a ‘traffic jam’ in the posterior tailbud and a severely contorted trunk, whereas decreases in both coherence and flow rate merely ‘kink’ the tip of the tail. Thus, the balance between flow rate and the coherence of collective migration within the tailbud steers zebrafish body elongation.
The tailbud is the posterior leading edge of the growing vertebrate embryo. Now, by measuring the three-dimensional cell flow field of the zebrafish tailbud, Scott Holley and co-workers reveal a posterior flow within the tailbud that reflects ordered collective cell migration (p. 573). They identify a transition in tissue fluidity at the tailbud tip where there is a decrease in the coherence of the cell flow but no alterations of cell velocities. Inhibition of Wnt or Fgf signalling reduces the coherence of the flow, but affects trunk and tail extension differently. By using computer simulations to interpret these complex phenotypes, the researchers show that a decrease in the coherence of the flow combined with a normal flow rate leads to a ‘traffic jam’ in the posterior tailbud and a severely contorted trunk, whereas decreases in both coherence and flow rate merely ‘kink’ the tip of the tail. Thus, the balance between flow rate and the coherence of collective migration within the tailbud steers zebrafish body elongation.
PLUS…
Transcriptional repressors: multifaceted regulators of gene expression
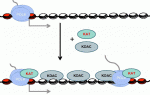 Although classic repressors undoubtedly silence transcription, genome-wide studies show that many repressors are associated with actively transcribed loci. Reynolds, O’Shaughnessy and Hendrich review this evidence and propose that the modulation of gene expression by co-repressor complexes provides transcriptional fine-tuning that drives development. See the Review on p. 505
Although classic repressors undoubtedly silence transcription, genome-wide studies show that many repressors are associated with actively transcribed loci. Reynolds, O’Shaughnessy and Hendrich review this evidence and propose that the modulation of gene expression by co-repressor complexes provides transcriptional fine-tuning that drives development. See the Review on p. 505
Establishing and maintaining gene expression patterns: insights from sensory receptor patterning
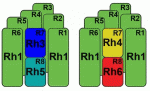 Rister, Desplan and Vasiliauskas review the mechanisms that generate and maintain sensory receptor expression patterns and compare them to those that control sensory receptor expression patterns in the mouse retina and in the mouse and fly olfactory systems. See the Primer article on p. 493
Rister, Desplan and Vasiliauskas review the mechanisms that generate and maintain sensory receptor expression patterns and compare them to those that control sensory receptor expression patterns in the mouse retina and in the mouse and fly olfactory systems. See the Primer article on p. 493
Click here to see other articles in the ‘Development: The Big Picture’ series.
Radial glia – from boring cables to stem cell stars
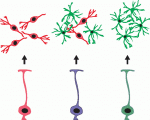 As part of the ‘Development Classics’ series, Malatesta and Gotz look back at their 2000 Development paper, in which they showed that radial glial cells act as neural stem and progenitor cells in development – a discovery that has led to a change in the concept of neural stem cells in the adult brain. See the Spotlight on p. 483
As part of the ‘Development Classics’ series, Malatesta and Gotz look back at their 2000 Development paper, in which they showed that radial glial cells act as neural stem and progenitor cells in development – a discovery that has led to a change in the concept of neural stem cells in the adult brain. See the Spotlight on p. 483
A germline-centric view of cell fate commitment, reprogramming and immortality
 At the recent EMBO/EMBL symposium ‘Germline – Immortality through Totipotency’, researchers discussed the mechanisms that establish and control totipotency, with an eye towards the mechanisms that may endow germ cells with the ability to propagate totipotency across generations. See the Meeting Report by Torres-Padilla and Ciosk on p. 487
At the recent EMBO/EMBL symposium ‘Germline – Immortality through Totipotency’, researchers discussed the mechanisms that establish and control totipotency, with an eye towards the mechanisms that may endow germ cells with the ability to propagate totipotency across generations. See the Meeting Report by Torres-Padilla and Ciosk on p. 487
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (5 votes)
(5 votes)
 We haven’t posted one of these in a while, but there are quite a few things coming up. Remember to also keep an eye on the
We haven’t posted one of these in a while, but there are quite a few things coming up. Remember to also keep an eye on the 
 The directed differentiation of pluripotent stem cells into endodermal derivatives, including insulin-producing pancreatic β cells, has considerable clinical promise in cell replacement therapies. The first step in this process is the conversion of pluripotent stem cells into definitive endoderm (DE). Here (p.
The directed differentiation of pluripotent stem cells into endodermal derivatives, including insulin-producing pancreatic β cells, has considerable clinical promise in cell replacement therapies. The first step in this process is the conversion of pluripotent stem cells into definitive endoderm (DE). Here (p.  Congenital biliary atresia is an incurable disease of newborn infants that is characterised by deformation of the gallbladder and biliary duct system. Yoshiakira Kanai and co-workers now report (p.
Congenital biliary atresia is an incurable disease of newborn infants that is characterised by deformation of the gallbladder and biliary duct system. Yoshiakira Kanai and co-workers now report (p.  During embryogenesis and gametogenesis, the DNA methyltransferases Dnmt3A and Dnmt3B establish the genome-wide methylation patterns that are essential for mammalian development and reproduction. The catalytically inert Dnmt3-like (Dnmt3L) is known to regulate de novo methylation in the germline but does it function in the early embryo? Déborah Bourc’his and colleagues have been investigating this question and, on p.
During embryogenesis and gametogenesis, the DNA methyltransferases Dnmt3A and Dnmt3B establish the genome-wide methylation patterns that are essential for mammalian development and reproduction. The catalytically inert Dnmt3-like (Dnmt3L) is known to regulate de novo methylation in the germline but does it function in the early embryo? Déborah Bourc’his and colleagues have been investigating this question and, on p.  Human hearts do not regenerate after a heart attack because adult mammalian cardiomyocytes proliferate poorly in response to injury. By contrast, zebrafish regenerate heart muscle after trauma by inducing cardiomyocyte proliferation. Studies of zebrafish heart regeneration might, therefore, identify ways to repair damaged human hearts. Here (p.
Human hearts do not regenerate after a heart attack because adult mammalian cardiomyocytes proliferate poorly in response to injury. By contrast, zebrafish regenerate heart muscle after trauma by inducing cardiomyocyte proliferation. Studies of zebrafish heart regeneration might, therefore, identify ways to repair damaged human hearts. Here (p.  During retinal development, seven retinal cell types are specified from a common pool of retina progenitor cells (RPCs). Several proneural basic helix-loop-helix (bHLH) transcriptional regulators, including Atoh7 and Neurod1, direct the intrinsic programming of RPCs but how do individual bHLH factors influence RPC fate? On p.
During retinal development, seven retinal cell types are specified from a common pool of retina progenitor cells (RPCs). Several proneural basic helix-loop-helix (bHLH) transcriptional regulators, including Atoh7 and Neurod1, direct the intrinsic programming of RPCs but how do individual bHLH factors influence RPC fate? On p.  The tailbud is the posterior leading edge of the growing vertebrate embryo. Now, by measuring the three-dimensional cell flow field of the zebrafish tailbud, Scott Holley and co-workers reveal a posterior flow within the tailbud that reflects ordered collective cell migration (p.
The tailbud is the posterior leading edge of the growing vertebrate embryo. Now, by measuring the three-dimensional cell flow field of the zebrafish tailbud, Scott Holley and co-workers reveal a posterior flow within the tailbud that reflects ordered collective cell migration (p.  Although classic repressors undoubtedly silence transcription, genome-wide studies show that many repressors are associated with actively transcribed loci. Reynolds, O’Shaughnessy and Hendrich review this evidence and propose that the modulation of gene expression by co-repressor complexes provides transcriptional fine-tuning that drives development. See the Review on p.
Although classic repressors undoubtedly silence transcription, genome-wide studies show that many repressors are associated with actively transcribed loci. Reynolds, O’Shaughnessy and Hendrich review this evidence and propose that the modulation of gene expression by co-repressor complexes provides transcriptional fine-tuning that drives development. See the Review on p.  Rister, Desplan and Vasiliauskas review the mechanisms that generate and maintain sensory receptor expression patterns and compare them to those that control sensory receptor expression patterns in the mouse retina and in the mouse and fly olfactory systems. See the Primer article on p.
Rister, Desplan and Vasiliauskas review the mechanisms that generate and maintain sensory receptor expression patterns and compare them to those that control sensory receptor expression patterns in the mouse retina and in the mouse and fly olfactory systems. See the Primer article on p.  As part of the ‘Development Classics’ series, Malatesta and Gotz look back at their 2000 Development paper, in which they showed that radial glial cells act as neural stem and progenitor cells in development – a discovery that has led to a change in the concept of neural stem cells in the adult brain. See the Spotlight on p.
As part of the ‘Development Classics’ series, Malatesta and Gotz look back at their 2000 Development paper, in which they showed that radial glial cells act as neural stem and progenitor cells in development – a discovery that has led to a change in the concept of neural stem cells in the adult brain. See the Spotlight on p.  At the recent EMBO/EMBL symposium ‘Germline – Immortality through Totipotency’, researchers discussed the mechanisms that establish and control totipotency, with an eye towards the mechanisms that may endow germ cells with the ability to propagate totipotency across generations. See the Meeting Report by Torres-Padilla and Ciosk on p.
At the recent EMBO/EMBL symposium ‘Germline – Immortality through Totipotency’, researchers discussed the mechanisms that establish and control totipotency, with an eye towards the mechanisms that may endow germ cells with the ability to propagate totipotency across generations. See the Meeting Report by Torres-Padilla and Ciosk on p.