Polarizing morphogenesis: epithelia dance the polonaise
Posted by Danelle Devenport, on 13 June 2018
The story behind our recent paper: Counter-rotational cell flows drive morphological and cell fate asymmetries in mammalian hair follicles. Maureen Cetera, Liliya Leybova, Bradley Joyce & Danelle Devenport, Nature Cell Biology.
Planar cell polarity (PCP) is a fascinating biological problem because it spans such vast biological scales, from nanometers to meters of spatial organization. When we think of PCP – the coordinated polarization of cellular structures across a tissue plane – we often imagine the Drosophila wing, where individual cells produce actin-rich protrusions that point in a uniform orientation. But PCP is also observed in much more complex epithelia like the vertebrate epidermis, where multicellular structures, which produce scales, feathers, or fur, collectively align across the entire skin surface. Although we showed nearly 10 years ago that mammalian hair follicles polarize through the same conserved core PCP pathway that orients Drosophila wing hairs (Devenport and Fuchs, 2008), how these multicellular structures polarize was an intractable question because we lacked the long-term live imaging methods to follow their morphogenesis at cellular resolution.
Movie 1
Enter Team Hair Follicle – a collaboration between two postdocs, Maureen Cetera and Brad Joyce, and a graduate student Liliya Leybova. Through trial and error they figured out that by culturing the embryonic epidermis at an air liquid interface, using a transgenic line expressing bright fluorescent membrane labels (mT/mG) and imaging with a spinning disc confocal to minimize phototoxicity, they could acquire time-lapse movies spanning 24 hours of epidermal development with cellular resolution. The first batch of movies took our breaths away. Never had I dreamed the developing skin would be so dynamic. We never imagined that skin epithelial cells, which we often think of a rigidly adherent with adherens junctions and desmosomes, would behave as if they were fluid and extensively rearrange (Movie 1).
Focusing on a 10-12 hour window of time during which hair placodes polarize, and using automated segmentation and cell tracking, we discovered that during polarization the placode epithelium undergoes extensive rearrangements organized in a counter-rotating pattern (Movie 2-3).
Movie 2
Movie 3
Cells that are positioned centrally move anteriorly to occupy the growing tip of the follicle while more peripheral cells are swept posteriorly and incorporate into the trailing rear (Figure 1).
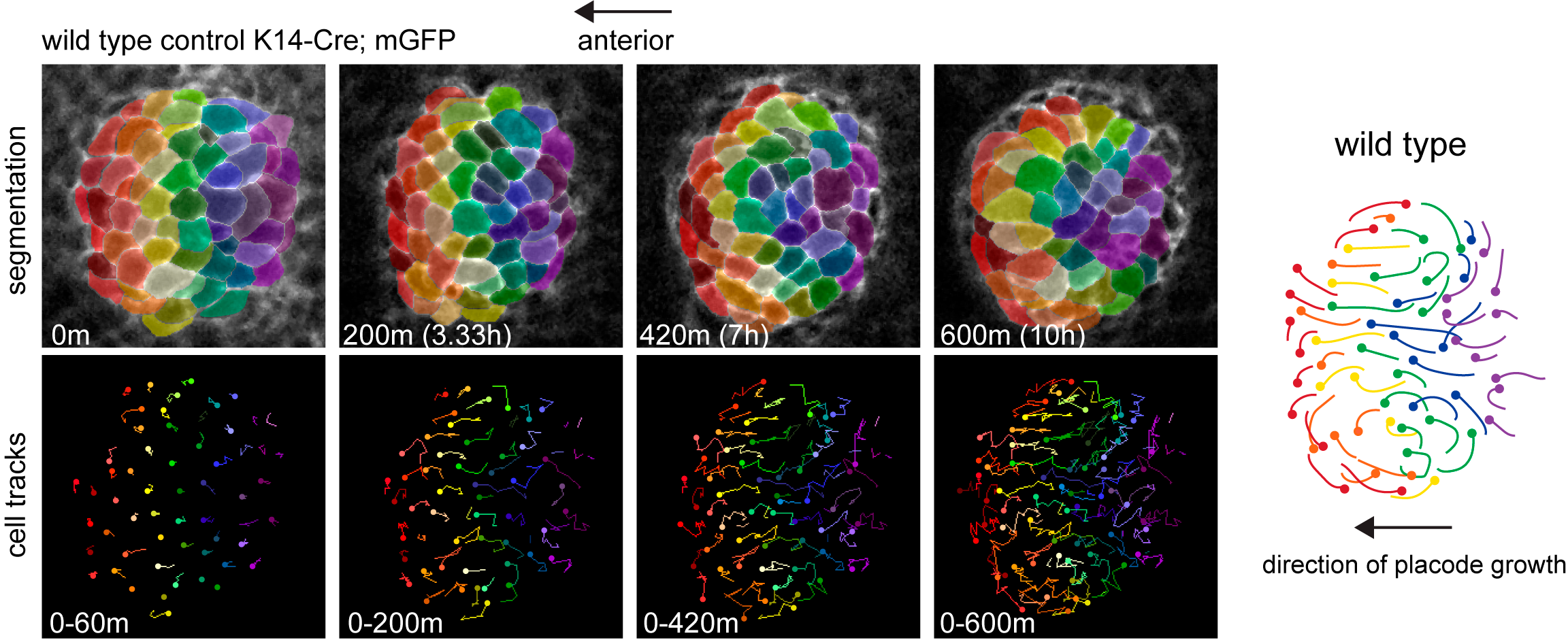
Figure 1
These movements were abolished in the absence of PCP, non-muscle myosin and Rho kinase activities, with the resulting follicles growing vertically rather than anteriorly (Figure 2, Movie 4).
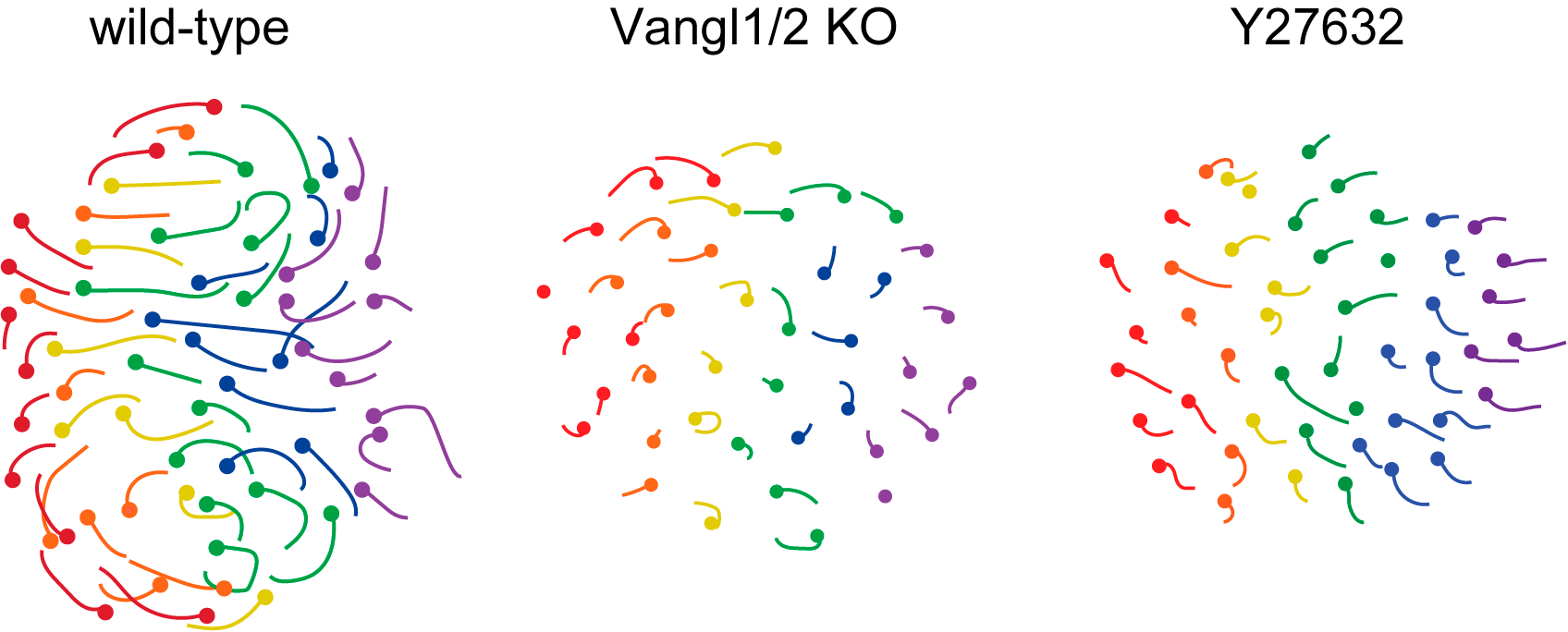
Figure 2
Movie 4
The counter-rotational movements we observed were striking because 1) they explained how the placode gains its morphological asymmetry, by completely remodeling the placode epithelium from a radial to planar polarized organization; and 2) the pattern of movements closely resemble the elaborate ‘polonaise’ movements of gastrulating chick embryos, a process previously linked to the PCP pathway (Voiculescu et al, 2007). During formation of the chick primitive streak, convergent extension in the posterior hemisphere of the embryo displaces cells in the anterior into two counter-rotating flows (Chuai and Weijer, 2008; Rozbicki et al, 2015). It is remarkable that the same pattern of cell movements involving thousands of cells in the chick embryo is made by just a few dozen cells confined within and scattered in a periodic pattern across the skin epithelium.
We were then confronted with the difficult task of connecting PCP to the counter-rotating rearrangements we observed. A key feature of the PCP pathway is that its core components, as well as downstream cytoskeletal factors, are asymmetrically localized to intercellular junctions. What was difficult to explain, and will require much more work to fully understand, was how the asymmetry of the core PCP components, which align along a common axis amongst all cells across the skin epithelium, could generate cell movements in opposite directions. So Maureen broke down the collective movement into its local component parts and asked how the asymmetry of PCP components correlated to those behaviors. First, she determined that in the posterior half of the placode, cells intercalate towards the placode midline via polarized shrinking and growth of intercellular junctions (Figure 3). Junctions that were lost tended to be vertically oriented where new junctions formed in a horizontal orientation. In the anterior half, cells also underwent neighbor exchanges but with the opposite polarity so they moved away from the midline and posteriorly (Figure 3).
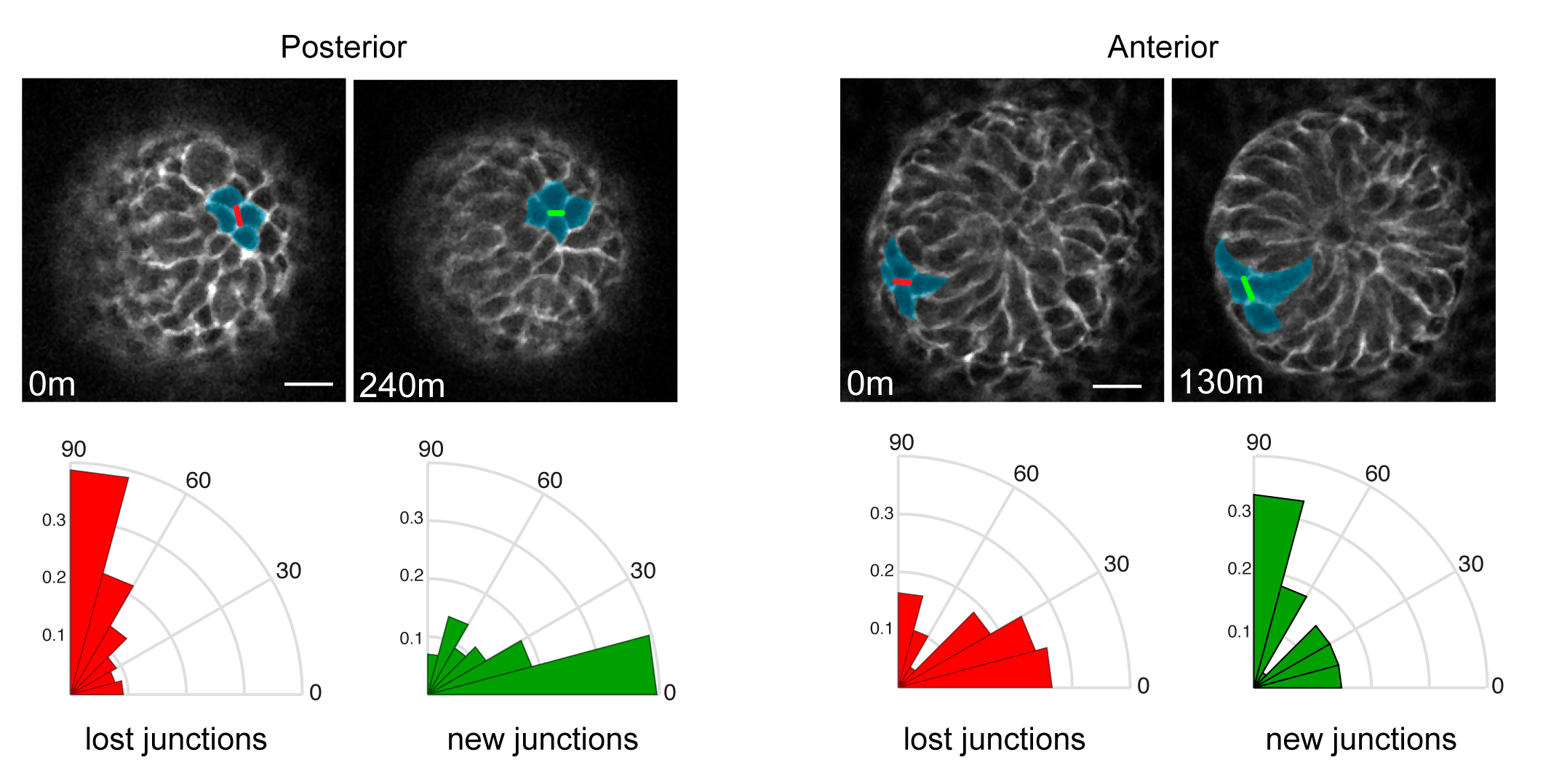
Figure 3
PCP protein localization correlated with junction shrinkage, even in the anterior half of the placode, where PCP junctions were rotated relative to the AP axis (Figure 4). This reminded us of a result from a previous study where we showed that when cells exchange neighbors through junction remodeling, the local axis of PCP asymmetry rotates relative to its original orientation (Aw et al, 2016). We hypothesized that early remodeling events in the posterior of the placode could cause PCP-enriched junctions to rotate, thereby generating a new PCP axis for junction shrinkage in the anterior.
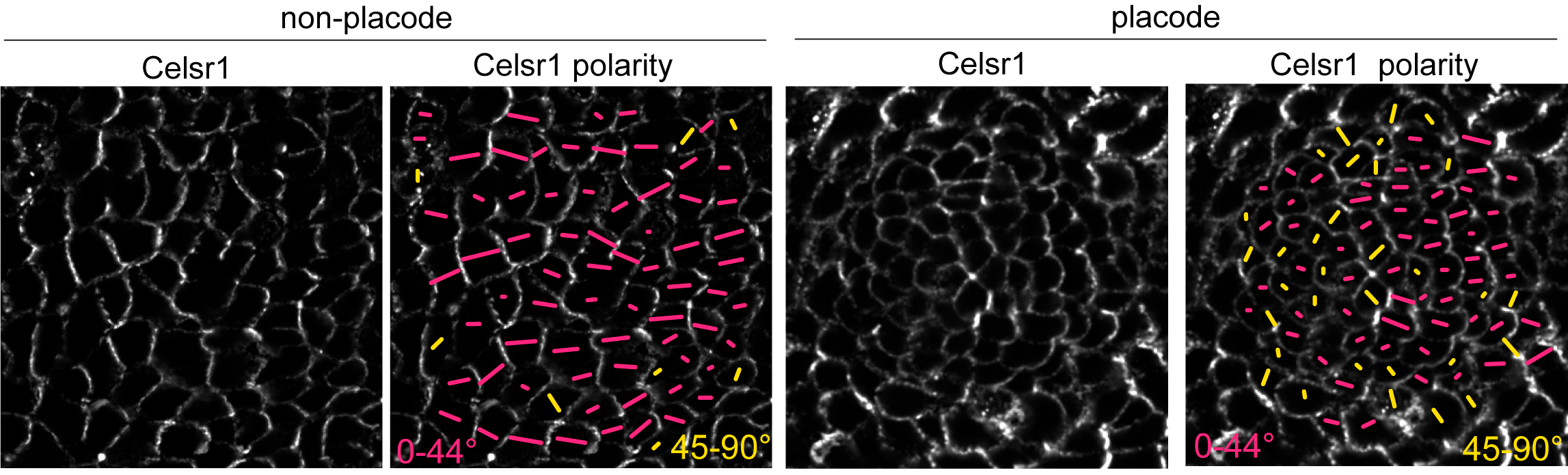
Figure 4
The discovery of counter-rotational movements in the placode also explained how PCP generates cell fate asymmetry. In our initial 2008 study we reported that the earliest two hair follicle lineages were distributed in a planar polarized organization (Devenport and Fuchs, 2008), with hair matrix precursors at the anterior and stem cell precursors at the posterior. But the relationship between morphological and cell fate asymmetry was unclear in part because the two events couldn’t be temporally separated. By live imaging we could now show that cell fate asymmetry arises from counter-rotating cell rearrangements. Initially, the two progenitor populations are specified in a bullseye pattern, with a central cluster of matrix progenitors surrounded by a halo of stem cell precursors. Counter-rotating cell rearrangements moved the two cell populations into their respective, anterior-posterior positions and shifted the placode from vertical to anterior-directed growth (Figure 5).
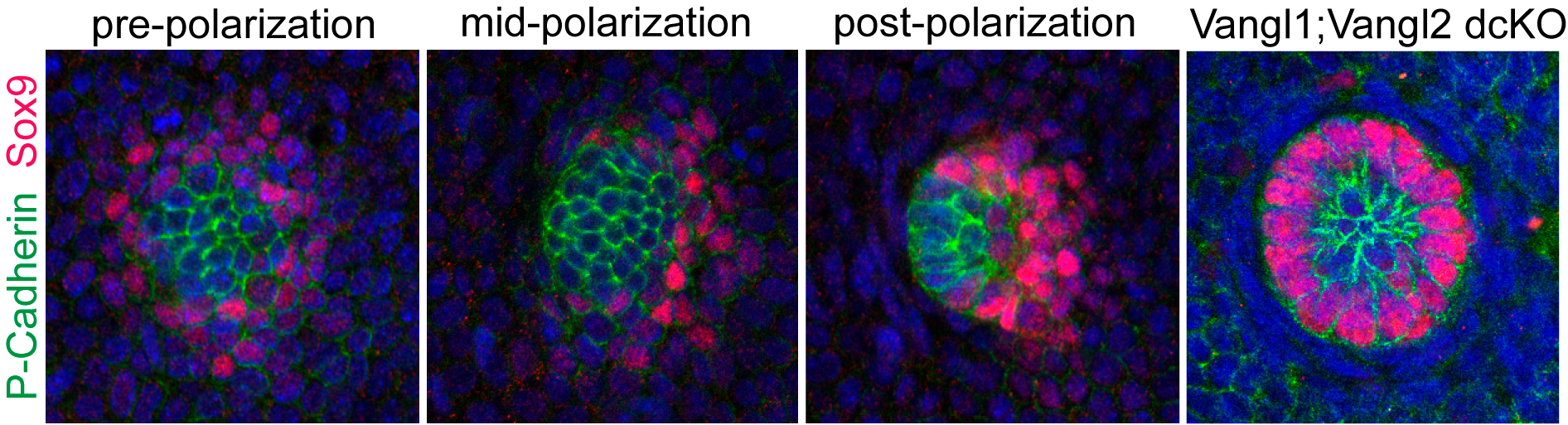
Figure 5
From these studies we learned that a just a single PCP-dependent process – counter rotating cell rearrangements – could generate both morphological and cell fate asymmetry. Additionally, we learned that PCP-dependent cell intercalation is a deeply conserved morphogenetic tool, used to shape tissues in ways other than convergent extension. We learned, of course, that live imaging reveals unexpected and delightful new insights beyond any hypothesis I was ever able to muster. This study was also an really successful experiment in team research. Whereas most projects in the lab and in my previous training had centered mostly around individuals, this was a true collaborative effort. Perhaps it is appropriate that the insights we gained about how PCP polarizes cell collectives would finally come, not from the efforts of just one individual, but from the collective.
References
Devenport, D. & Fuchs, E. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol 10, 1257-1268, doi:10.1038/ncb1784 (2008).
Voiculescu, O., Bertocchini, F., Wolpert, L., Keller, R. E. & Stern, C. D. The amniote primitive streak is defined by epithelial cell intercalation before gastrulation. Nature 449, 1049-1052, doi:10.1038/nature06211 (2007).
Chuai, M. & Weijer, C. J. The mechanisms underlying primitive streak formation in the chick embryo. Curr Top Dev Biol 81, 135-156, doi:10.1016/S0070-2153(07)81004-0 (2008).
Rozbicki, E. et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat Cell Biol 17, 397-408, doi:10.1038/ncb3138 (2015).
Aw, W. Y., Heck, B. W., Joyce, B. & Devenport, D. Transient Tissue-Scale Deformation Coordinates Alignment of Planar Cell Polarity Junctions in the Mammalian Skin. Curr Biol 26, 2090-2100, doi:10.1016/j.cub.2016.06.030 (2016).


 (No Ratings Yet)
(No Ratings Yet) (2 votes)
(2 votes)