Postdoctoral position in vascular development using spatial transcriptomics, tissue clearing, and light-sheet microscopy
Posted by Douglas Shepherd, on 31 October 2017
Closing Date: 15 March 2021
|
Posted by Douglas Shepherd, on 31 October 2017
Closing Date: 15 March 2021
|
Posted by BSDB, on 31 October 2017
![]() Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Our third report from the 2017 group of student awardees comes from Rachael Adams (student at University of Cambridge), who undertook her studentship with Peter Lawrence at the Dept. of Zoology in Cambridge.
This summer, thanks to the Gurdon/The Company of Biologists Summer Studentship, I was fortunate to spend 8 weeks working in Dr Peter Lawrence’s lab in the Department of Zoology.
The group studies PCP, a pathway which coordinates cell polarity and helps to align epidermal patterns in the Drosophila abdomen. Drosophila larvae are covered with a cuticle decorated by denticles which help the larvae to grip substrate in order to move. These denticles form a specific pattern and changes in the pattern can be used to investigate the properties of the PCP pathway. PCP genes are highly conserved and have been identified as being involved in processes such as vertebrate gastrulation, demonstrating their fundamental importance in animal development and patterning.
My project aimed to investigate the importance of Rab GTPases to PCP. Much research has been carried out on the Rab family of proteins, as they act as master regulators of intracellular membrane trafficking. The accurate delivery of cargo between organelles is crucial for normal eukaryotic cell function. Rabs facilitate this by coordinating vesicle formation, transport and fusion.
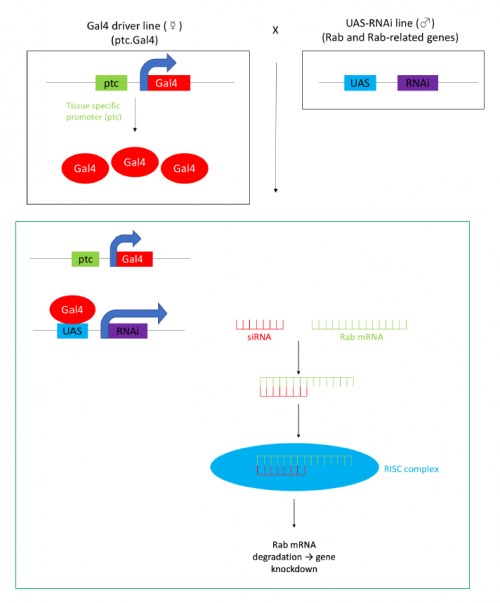
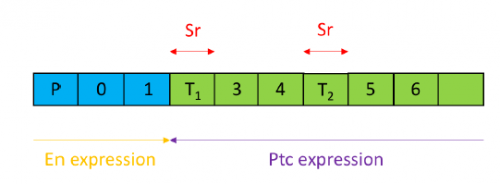
In order to test the role of as many Drosophila Rabs as possible I carried out a UAS RNAi knockdown screen of 34 different Rab and Rab-related genes, see figure 1. This screen took 7 weeks to carry out as I had to cross a collection of 103 different UAS-RNAi lines with ptc.Gal4 virgin females. This involved maintaining a population of ptc.Gal4 flies in several bottles. I crossed the virgins with males obtained from each UAS-RNAi stock and incubated the flies at 29℃, the optimal temperature to see the effect of the RNAi. Once each cross was made it was approximately 5 days before third instar larvae could be collected. To identify any change in the phenotype, I mounted several third instar larval progeny of each Gal4-UAS- RNAi cross and examined the denticle pattern produced under the microscope. Most larvae exhibited a wild type pattern, this may be because most of the Rabs I examined don’t have a function in PCP or redundancy of Rab proteins may mask certain phenotypes. The low efficacy of some of the RNAi constructs may have also affected the proportion of successful knockdowns. However, I found that several of the Rab23 crosses produced an unusual phenotype where the tendon cell gaps appeared larger than normal, see figure 2. In order to test whether this result is repeatable, I have crossed the UAS-RNAi Rab23 stocks to different driver lines (sr.Gal4 and en.Gal4). As Gal4 expression is under the control of a different promoter in each driver line, this results in a different expression pattern of the RNAi, see figure 3. Whilst different drivers will not produce the same phenotype, if the denticle pattern is disrupted it may help in the investigation of the role of Rab23 in PCP.
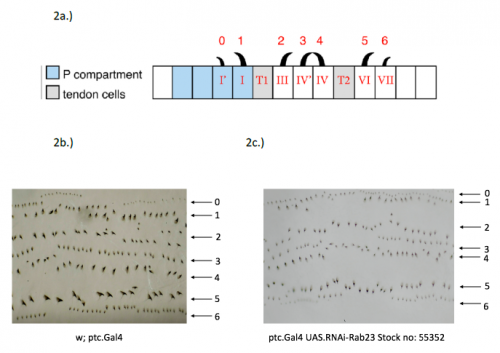
The results from the ptc.Gal4- UAS-RNAi Rab 23 crosses raised the question of why the tendon cell gap appeared larger. Possibilities include: that the tendon cells were enlarged, divided more frequently or the surrounding cells were smaller. To investigate this phenotype, I set up a series of crosses and aim to ultimately generate larvae carrying the Rab23 knockdown and expressing spaghetti squash protein (sqh) labelled with the fluorescent marker mCherry. Sqh encodes the regulatory light chain of non-muscle myosin so is expressed throughout cells. The sqh.mCherry construct therefore allows individual cells to be visualised with a confocal microscope and could help identify why the tendon cell gap is expanded.
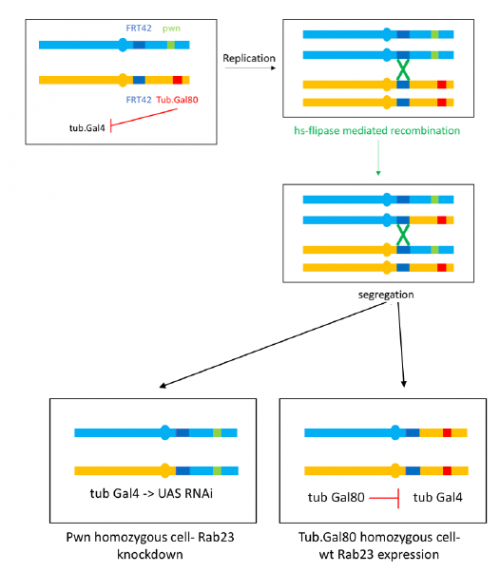
To further investigate the phenotype I set up crosses to produce marked clones in adult flies where Rab23 is knocked down, the resultant mosaic animal allows comparison with wild type cells. This is achieved by using the Flp-FRT system to bring about site-specific recombination, see figure 4.
I have really enjoyed my time in the lab and have learned a great deal. I am looking forward to continuing to investigate the Rab23 phenotype and seeing the results of my crosses as part of my Part II Zoology project this term. My special thanks to José Casal for supervising and to everyone in the lab for providing a great deal of help and advice.
Posted by Seema Grewal, on 31 October 2017
Here are the highlights form the current issue of Development:
 The tectorial membrane (TM) is an extracellular matrix (ECM) that overlies the organ of Corti in the inner ear and is crucial for our sense of hearing. It is composed of collagen fibrils embedded in a tectorin-based matrix. The precise alignment of the collagen fibrils across the TM is a feature considered critical for hearing, but very little is known about how this pattern is generated. On p. 3978, Richard Goodyear and colleagues undertake a detailed analysis of TM development in mice and begin to investigate the mechanisms underlying collagen-fibril orientation. They find that the presence of a tectorin-based matrix is essential for the normal co-alignment and orientation of the first-forming collagen fibrils, and that collagen-fibril orientation does not seem to depend on stretch of the ECM caused by growth of the underlying epithelium. Rather, the authors identify an influence of the planar cell polarity machinery, generally associated with cell-cell alignment, on collagen fibril orientation – although the molecular mechanisms underlying this remain unclear. These data provide first insights into how TM patterning is achieved, and point to an intriguing interplay between planar cell polarity and collagen-fibril organisation.
The tectorial membrane (TM) is an extracellular matrix (ECM) that overlies the organ of Corti in the inner ear and is crucial for our sense of hearing. It is composed of collagen fibrils embedded in a tectorin-based matrix. The precise alignment of the collagen fibrils across the TM is a feature considered critical for hearing, but very little is known about how this pattern is generated. On p. 3978, Richard Goodyear and colleagues undertake a detailed analysis of TM development in mice and begin to investigate the mechanisms underlying collagen-fibril orientation. They find that the presence of a tectorin-based matrix is essential for the normal co-alignment and orientation of the first-forming collagen fibrils, and that collagen-fibril orientation does not seem to depend on stretch of the ECM caused by growth of the underlying epithelium. Rather, the authors identify an influence of the planar cell polarity machinery, generally associated with cell-cell alignment, on collagen fibril orientation – although the molecular mechanisms underlying this remain unclear. These data provide first insights into how TM patterning is achieved, and point to an intriguing interplay between planar cell polarity and collagen-fibril organisation.
 Embryonic patterning is dependent on the establishment of the anteroposterior (AP) and dorsoventral axes early in development. In mammalian embryos, these axes are established by a breaking of symmetry in the epiblast, which involves signals from the extra-embryonic tissues. However, the molecular mechanisms that control this process are still not fully understood. On p. 3894, David Turner, Alfonso Martinez Arias and colleagues use gastruloids, three-dimensional aggregates of mouse embryonic stem cells, as a tool to unravel the signalling pathways that establish AP polarity in mammalian embryos. The authors demonstrate that these gastruloids can develop an AP axis in the absence of extra-embryonic tissue, instead depending on precisely timed interactions between Wnt and Nodal signalling. They also show that BMP signalling is dispensable for AP axis formation. This research demonstrates the powerful potential of gastruloids as a tool to understand the molecular mechanisms that underpin early embryonic development. Together, their results suggest that extra-embryonic tissues do not induce axis formation per se, but rather bias the critical symmetry-breaking event in embryo development, furthering our understanding of the molecular control of embryonic patterning.
Embryonic patterning is dependent on the establishment of the anteroposterior (AP) and dorsoventral axes early in development. In mammalian embryos, these axes are established by a breaking of symmetry in the epiblast, which involves signals from the extra-embryonic tissues. However, the molecular mechanisms that control this process are still not fully understood. On p. 3894, David Turner, Alfonso Martinez Arias and colleagues use gastruloids, three-dimensional aggregates of mouse embryonic stem cells, as a tool to unravel the signalling pathways that establish AP polarity in mammalian embryos. The authors demonstrate that these gastruloids can develop an AP axis in the absence of extra-embryonic tissue, instead depending on precisely timed interactions between Wnt and Nodal signalling. They also show that BMP signalling is dispensable for AP axis formation. This research demonstrates the powerful potential of gastruloids as a tool to understand the molecular mechanisms that underpin early embryonic development. Together, their results suggest that extra-embryonic tissues do not induce axis formation per se, but rather bias the critical symmetry-breaking event in embryo development, furthering our understanding of the molecular control of embryonic patterning.
 The core autophagy protein Atg16L1 has been identified as a genetic risk factor in inflammatory bowel disease, but how it plays this role has remained unclear. On p. 3990, Gábor Juhász and colleagues interrogate the role of Atg16, the Drosophila orthologue of human ATG16L1, in intestinal homeostasis and inflammation. Using mutants that affect either the N-terminal autophagic domain or the C-terminal WD40 domain, they observe defects in intestinal morphology and an impaired stress response in Atg16 WD40 mutants. In Atg16 WD40 mutant intestines, the differentiation of enteroendocrine (EE) cells is impaired, leading to an accumulation of pre-EE cells, and this results from reduced Slit/Robo signalling (a pathway known to regulate EE cell number). The failure of EE differentiation is accompanied by an inflammatory response, but appears to be independent of autophagy: autophagy is not altered in Atg16 WD40 mutants, and mutants affecting the autophagy domain alter neither Slit/Robo signalling nor EE differentiation. Finally, the authors show that Atg16 binds to the GTPase Rab19 – also a genetic risk factor for inflammatory bowel disease – and the two cooperate in regulating intestinal homeostasis. This work provides insight into the molecular control of intestinal homeostasis and implies a link between impaired cell differentiation and intestinal pathologies in humans.
The core autophagy protein Atg16L1 has been identified as a genetic risk factor in inflammatory bowel disease, but how it plays this role has remained unclear. On p. 3990, Gábor Juhász and colleagues interrogate the role of Atg16, the Drosophila orthologue of human ATG16L1, in intestinal homeostasis and inflammation. Using mutants that affect either the N-terminal autophagic domain or the C-terminal WD40 domain, they observe defects in intestinal morphology and an impaired stress response in Atg16 WD40 mutants. In Atg16 WD40 mutant intestines, the differentiation of enteroendocrine (EE) cells is impaired, leading to an accumulation of pre-EE cells, and this results from reduced Slit/Robo signalling (a pathway known to regulate EE cell number). The failure of EE differentiation is accompanied by an inflammatory response, but appears to be independent of autophagy: autophagy is not altered in Atg16 WD40 mutants, and mutants affecting the autophagy domain alter neither Slit/Robo signalling nor EE differentiation. Finally, the authors show that Atg16 binds to the GTPase Rab19 – also a genetic risk factor for inflammatory bowel disease – and the two cooperate in regulating intestinal homeostasis. This work provides insight into the molecular control of intestinal homeostasis and implies a link between impaired cell differentiation and intestinal pathologies in humans.
PLUS:
 Christiane Nüsslein-Volhard is Director Emeritus at the Max Planck Institute for Developmental Biology in Tübingen, Germany. In 1995, she was awarded the Nobel Prize for Physiology and Medicine, along with Eric Wieschaus and Edward Lewis, for her work on the genetic control of embryogenesis using the fruit fly Drosophila melanogaster. In the 1990s, she transitioned her lab to working with zebrafish (Danio rerio), using similar forward genetic approaches to those that had proved so successful in Drosophila to uncover key regulators of vertebrate development. We met with Christiane at the recent International Society for Developmental Biology (ISDB) meeting in Singapore, to talk about her research, the impact of the Nobel Prize and the challenges of being a ‘woman in science’. See the Spotlight article.
Christiane Nüsslein-Volhard is Director Emeritus at the Max Planck Institute for Developmental Biology in Tübingen, Germany. In 1995, she was awarded the Nobel Prize for Physiology and Medicine, along with Eric Wieschaus and Edward Lewis, for her work on the genetic control of embryogenesis using the fruit fly Drosophila melanogaster. In the 1990s, she transitioned her lab to working with zebrafish (Danio rerio), using similar forward genetic approaches to those that had proved so successful in Drosophila to uncover key regulators of vertebrate development. We met with Christiane at the recent International Society for Developmental Biology (ISDB) meeting in Singapore, to talk about her research, the impact of the Nobel Prize and the challenges of being a ‘woman in science’. See the Spotlight article.
 During development, genes are transcribed at specific times, locations and levels. In recent years, the emergence of quantitative tools has significantly advanced our ability to measure transcription with high spatiotemporal resolution in vivo. Here, Angela DePace and co-workers highlight recent studies that have used these tools to characterize transcription during development, and discuss the mechanisms that contribute to the precision and accuracy of the timing, location and level of transcription. See the Review.
During development, genes are transcribed at specific times, locations and levels. In recent years, the emergence of quantitative tools has significantly advanced our ability to measure transcription with high spatiotemporal resolution in vivo. Here, Angela DePace and co-workers highlight recent studies that have used these tools to characterize transcription during development, and discuss the mechanisms that contribute to the precision and accuracy of the timing, location and level of transcription. See the Review.
 Cortical interneurons are a diverse group of neurons that project locally and are crucial for regulating information processing and flow throughout the cortex. Recent studies in mice have advanced our understanding of how these neurons are specified, migrate and mature. Here, John Rubenstein and colleagues evaluate new findings that provide insights into the development of cortical interneurons and that shed light on when their fate is determined, on the influence that regional domains have on their development, and on the role that key transcription factors and other crucial regulatory genes play in these events. See the Review.
Cortical interneurons are a diverse group of neurons that project locally and are crucial for regulating information processing and flow throughout the cortex. Recent studies in mice have advanced our understanding of how these neurons are specified, migrate and mature. Here, John Rubenstein and colleagues evaluate new findings that provide insights into the development of cortical interneurons and that shed light on when their fate is determined, on the influence that regional domains have on their development, and on the role that key transcription factors and other crucial regulatory genes play in these events. See the Review.
Posted by Kota Miura, on 31 October 2017
Dear Colleagues,
 The 2nd NEUBIAS school for Bioimage Analysts will be organized in Jan. 2018 in Szeged, Hungary, and the registration is now open (Organizers: Jean-Yves Tinevez & Kota Miura). Please visit the linked URL below for more details. This school is the most advanced among three levels of NEUBIAS school. Deadline for the registration is Nov. 9th.
The 2nd NEUBIAS school for Bioimage Analysts will be organized in Jan. 2018 in Szeged, Hungary, and the registration is now open (Organizers: Jean-Yves Tinevez & Kota Miura). Please visit the linked URL below for more details. This school is the most advanced among three levels of NEUBIAS school. Deadline for the registration is Nov. 9th.
The school is part of one week-conference, and there will be another school in parallel: “Early Career Investigator (ECI) school” In this school, you can learn how to program ImageJ macro and MATLAB script for bioimage analysis.
After these schools, there is 3-days symposium. Anyone can join and there is no selection for this meeting. School participants are free to attend this symposium.
Analyst School: http://goo.gl/qdWc6t
ECI school: http://goo.gl/Tfmwjx
Symposium: http://goo.gl/pHU3Pz
We are looking forward to seeing you in Szeged!
Sincerely,
Jean-Yves Tinevez and Kota Miura
Posted by Savraj Grewal, on 31 October 2017
Closing Date: 15 March 2021
The lab of Savraj Grewal (University of Calgary, Canada) is looking to recruit new postdocs and grad students.
Our lab investigates how growth is controlled during animal development. We use a combination of molecular and genetic approaches to investigate the cell-cell signalling pathways and the genetic mechanisms that govern the control of cell, tissue and body growth in Drosophila. Our main focus to-date has been the conserved insulin and TOR kinase pathways, and understanding how they regulate cellular and animal metabolism to drive growth. Further information on our research can be found here. Recent publications can be found here.
POSTDOCS: applicants with a Ph.D. and strong background in developmental biology, genetics, or molecular biology are encouraged to apply. Interested individuals should send a CV and a short statement of research interests to grewalss@ucalgary.
GRAD STUDENTS: applicants with a strong undergraduate degree in any area related to the biological sciences are encouraged to apply. Interested individuals should send a CV and a short statement of research interests to grewalss@ucalgary.
Posted by Kristin Gribble, on 30 October 2017
Closing Date: 15 March 2021
The Marine Biological Laboratory seeks a motivated Postdoctoral Scientist to join the laboratories of Kristin Gribble and David Mark Welch in the Josephine Bay Paul Center. The successful candidate will develop genome editing techniques, including CRISPR/Cas9, in rotifers, a novel aquatic invertebrate model system for studies of aging, neurobiology, developmental biology, ecology, and evolution. Specific goals of the project include designing guide RNAs, optimizing microinjection methodologies, phenotyping and genotyping mutant strains, and screening genes of interest.
Basic Qualifications:
Applicants should have a Ph.D. in biology, cell/molecular biology, biochemistry, or a related field. This position requires proficiency in basic molecular biology techniques, microscopy, microinjection, and CRISPR/Cas9 methodology. We are seeking an independent, organized, enthusiastic, and productive individual with robust problem solving skills. Excellent written, verbal and interpersonal skills, attention to detail, and a strong work ethic are essential. Position level and salary will depend upon education and experience.
Preferred Qualifications:
The ideal candidate will have working familiarity with RNAi techniques, transgenic protocols, and confocal microscopy. Proficiency in bioinformatics is a plus. Previous experience in established animal model or in non-model systems is preferred.
Special Instructions to Applicants:
Please submit the following three items with your application:
Please apply at: https://mbl.simplehire.com/postings/3824
Posted by BSDB, on 30 October 2017
When I tell my friends that I spent my summer looking down a microscope at worms they often give a snigger. My enthusiasm on the subject quickly earned me the affectionate (I hope) nickname, ‘Wormboy’. Yet the Wormboys and girls of Richard Poole’s lab instilled in me a great thirst for scientific study over the 8 weeks I spent with them, which I will certainly carry forward in my career. The lab studies the nematode worm C. elegans, and my job was to help characterise a glia-to-neuron transition they discovered in the male tail that occurs during sexual maturation. During my time, I learned how to handle worms, keep them healthy, perform crosses, use a fluorescent microscope and present my data to an audience. All of this as part of my first lab experience was not only invaluable, but very enjoyable, and I came away wanting more.
Worm death was a regular part of lab life. Saving them from starvation and disease was a constant uphill battle, one that at first I found insurmountable. Along with those natural causes of death, they also had to contend me, often crushing them clumsily under my pick and purging potentially contaminative stragglers on said pick in the Bunsen. Happily for the worms, such fates became less and less frequent, and I was eventually able to sustain healthy populations from which I selected young males to dunk in the intensely toxic chemical sodium azide, all the better to view them under fluorescent microscopy. A necessary sacrifice.
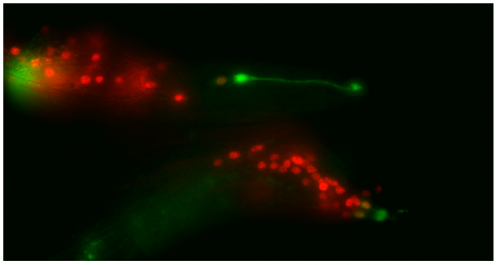
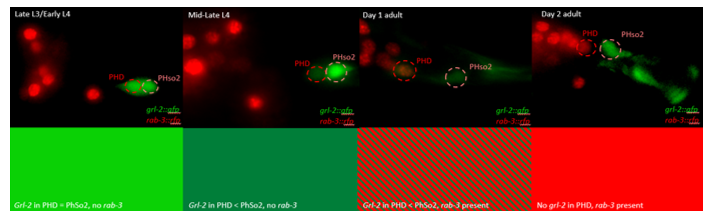
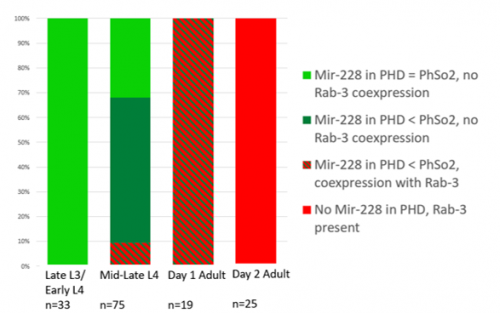
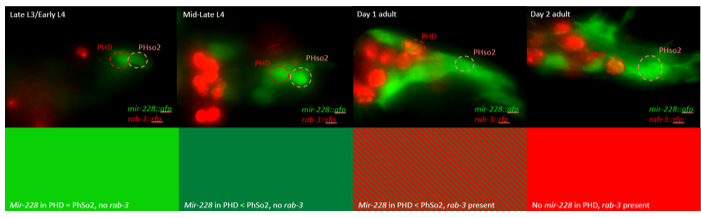
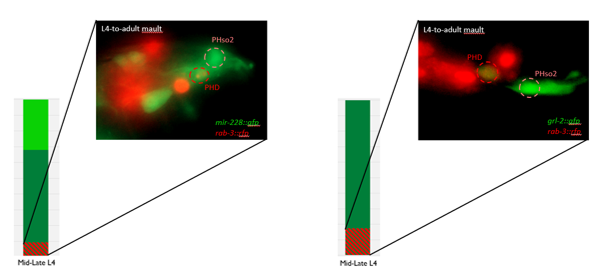
This microscopy work was the first step in characterising the glia-to-neuron transition of phasmid socket 1 (PhSo1) into the neuron PHD, which occurs in the male tail during sexual maturation. The lab had previously identified a similar transition of the amphid socket to MCM neuron in the head of the male (Sammut et al., 2015), though the PHD transition differs in how the neuron is formed. Where the amphid socket divides asymmetrically to generate the MCM, the PHD derives from PHso1 by direct trans-differentiation, without a division, while its sister- PHso2- remains glial. This new neuron’s activity was found to be linked to initiation of a novel behaviour employed by males during mating thought to improve their chance of spicule insertion. To track the changing identity of PHso1, I compared its expression of glial and neuronal markers linked to GFP and RFP respectively with that of PHso2. The two glial markers I used were grl-2 (socket specific protein) and mir-228 (pan-glial microRNA), and the neuronal marker a nuclear synaptic fusion protein, rab-3.
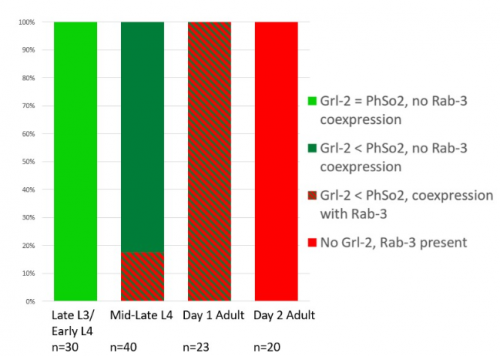
The glia-to-neuron transition appears to start in males of larval instar 4 (L4), while the gonad is taking shape. Through picking many, many worms, I showed a clear gradient of change of marker expression in PhSo1 compared with PhSo2:
Of course, fluorescent proteins do not perfectly match levels of the markers they represent, rather they give a general overview of the transformation. At the very least, this result shows PhSo1 might partially dedifferentiate before it eventually acquires neuronal characteristics when the worm becomes an adult. Understanding trans-differentiation events such as this is key to understanding how nature itself reassigns cell fate. And if you’re trying to do something yourself in cell biology, it often pays to learn how nature beat you to it. To determine the mechanics of this trans-differentiation would require a more quantitative technique: single molecule fluorescent in situ hybridisation (smFISH).
SmFISH employs fluorescently tagged RNA oligomers, antisense to an mRNA (or miRNA) of interest. Combined, these oligos fluoresce strongly enough that individual RNAs can be resolved and counted within PHso1. Tracking change in RNA quantity rather than GFP fluorescence would tell you whether or not glial expression stops before neuronal expression begins. Does the cell become completely naive? Perhaps only partially? Or maybe there’s no dedifferentiation at all, and the cell passes through a totally novel identity- part glia, part neuron. Though I didn’t have time to perform these experiments, I did create the necessary strains. By crossing a markerless strain with my GFP/RFP animals, and then selecting against RFP over a couple of generations I rendered worms expressing only one of the two GFP markers so that the RFP reporter used in smFISH would be visible.
Though the work I was doing represented a project in its early stages and was at times very laborious, I quickly got an appetite for it. There was a thrill to it, knowing that no one had ever done what I was doing. I felt that buzz again when I arranged the prettiest pictures I could muster into figures to show to the lab, and then again when they ceremoniously scoured my findings with the same level of scrutiny I had seen them exact on each other. Alongside my data, I showed off a particularly pretty picture of both the worm’s glia-to-neuron transitions occurring at once in the same animal, which I think will remain my crowning achievement in science for a long time (Richard even kindly offered to steal it for his presentations). The BSDB’s grant has allowed me a taste of what a career in academia can offer. Straightforward experiments fraught with unforeseen difficulties. Working on weekends when, infuriatingly, age-specific experiments simply weren’t possible on Friday. Enough money to live off (just). But above all, the enormous reward in discovering something new. And getting to work with some really, really great people. Thanks guys.
Sammut, M., Cook, S., Nguyen, K., Felton, T., Hall, D., Emmons, S., Poole, R. and Barrios, A. (2015). Glia-derived neurons are required for sex-specific learning in C. elegans. Nature, 526(7573), pp.385-390.
Posted by Julien Courchet, on 30 October 2017
Closing Date: 15 March 2021
A ERC funded postdoctoral position is available in the laboratory of Julien Courchet at the NeuroMyoGene Institute within the University of Lyon, France.
Our group studies the molecular mechanisms underlying axon outgrowth and neural circuits formation in the mouse cerebral cortex. Our current research is supported by an ERC Starting Grant and funds from AFM-telethon to explore how a dynamic regulation of the energy metabolism is involved in axon morphogenesis and cortex development. We focus on a previously identified kinase pathway controlling terminal axon branching through the regulation of mitochondria trafficking and distributing in developing axons (Cell 2013). Building on this previous research, the proposed project will use a combination of whole cell metabolomics, real-time fluorescent videomicroscopy and live 2-photon imaging to characterize some of the molecular mechanisms involved in the local regulation of mitochondria function in developing axon in vivo.
The selected candidate will join a young research team within a dynamic and collaborative scientific environment at the newly created NeuroMyoGene Institute (INMG). The candidate will have access to state-of-the-art facilities for imaging and metabolic analyses, including high quality confocal and 2-photon microscopes, animal phenotyping centers and a seahorse analyzer. Our institute is located in a newly renovated laboratory space in the Rockefeller faculty of Medicine in close proximity to the Neuroscience and the Cancer Research Centers.
Applicants should have a PhD degree or equivalent with a strong background and practical experience in neurobiology, confocal microscopy and/or real-time imaging. Previous experience working with rodent models is required. Training in techniques relevant to cell signaling, metabolic regulation and optogenetics would be an asset. We are looking for a highly motivated candidate with a strong attitude towards independent work and good interpersonal and communication skills. Excellent written and spoken English skills are essential. Ability to speak French in not mandatory.
The initial appointment is one year and can be renewed for 2 additional years. Salary including benefits will depend on previous experience according to guidelines at the University of Lyon. Applications will be reviewed on a rolling basis until position is filled. Selected candidates will be invited for an interview early 2018. Project start date is expected during the first semester of 2018.
Interested candidates should contact Dr Julien Courchet (julien.courchet@inserm.fr) with their CV, a summary of their previous research (< 1 page), a brief statement of their research interests and career goals, as well as the contact information for at least 3 references.
Posted by ecloke, on 30 October 2017
Closing Date: 15 March 2021

The MRC Weatherall Institute of Molecular Medicine (WIMM) has fully funded 4-year Prize PhD (DPhil) Studentships available to start in October 2018. These Studentships are open to outstanding students of any nationality who wish to train in experimental and/or computational biology.
The Institute is a world leading molecular and cell biology centre that focuses on research with application to human disease. It includes the recently opened MRC WIMM Centre for Computational Biology and houses over 500 research and support staff in 50 research groups working on a range of fields in Haematology, Gene Regulation & Epigenetics, Stem Cell Biology, Computational Biology, Cancer Biology, Human Genetics, Infection & Immunity. The Institute is committed to training the next generation of scientists in these fields through its Prize PhD Studentship Programme.
The fully funded studentships include a stipend of £18,000 per annum and cover University and College fees.
Further information on the studentships, how to apply, and the projects available can be found at:
http://www.imm.ox.ac.uk/wimm-prize-studentships-2018
Closing date for submission of applications: Monday, 8 January 2018, 12 noon UK time.
Interviews will take place the week commencing 22 January 2018.
Hashem Koohy – Machine-learning in gene function, transcription regulation and immunology
Ed Morrissey – Quantitative biology of cell fate
Aleksandr Sahakyan – Regulatory chromosomal domains and genome architecture
Supat Thongjuea – Computational biology of single-cell transcription and gene regulation
Ahmed Ahmed – Experimental therapeutics
Chris Babbs – Causes of congenital anaemia
Oliver Bannard – B cell biology
Andrew Blackford – DNA damage and disease
Walter Bodmer – Colorectal cancer, stem cells, differentiation & drug response
Marella De Bruijn – Developmental haematopoiesis
Zam Cader – Stem cell neurological disease models
Vincenzo Cerundolo – Tumour immunology, vaccine strategies
David Clynes – DNA damage, repair and cancer
Simon Davis – T-cell biology
Hal Drakesmith – Iron and infection
Christian Eggeling – Super-resolution microscopy in immunology
Ben Fairfax – Inflammation, genetics and cancer therapeutics
Marco Fritzsche – Biophysical immunology
Lars Fugger – Multiple sclerosis
Tudor Fulga – MicroRNAs in development and disease
Richard Gibbons – Chromatin, epigenetics & transcription
Anne Goriely – De novo mutations and human disease
Doug Higgs – Gene regulation and epigenetics
Ling-Pei Ho – Lung immunology
Georg Hollander – T cell development and thymus organogenesis
David Jackson – Lymphatic trafficking in inflammation and cancer
Peter McHugh – DNA repair
Adam Mead – Normal and leukaemic haematopoietic stem cell biology
Claus Nerlov – Tissue stem cell genetics
Graham Ogg – Translational skin research
Catherine Porcher – Transcription factors and blood development
Jan Rehwinkel – Innate detection of viruses
Irene Roberts – Trisomy 21, haematopoiesis and leukaemia
Tatjana Sauka-Spengler – Neural crest gene regulatory networks
Alison Simmons – Innate immunity & Crohn’s disease
Alain Townsend – Influenza and ebola, vaccination and treatment
Paresh Vyas – Leukaemic stem cells
Andrew Wilkie – Sperm and craniofacial mutations
![]()
Posted by Felipe Karam Teixeira, on 27 October 2017
Closing Date: 15 March 2021
The Karam Teixeira laboratory (https://www.gen.cam.ac.uk/research-groups/karam-teixeira) at the University of Cambridge (Department of Genetics) is looking to recruit an outstanding Postdoctoral scientist to investigate the molecular mechanisms sheltering totipotency and controlling germline stem cell behavior in vivo. Using the Drosophila germline as a model for studying stem cells, we employ an integrated approach, combining high-throughput molecular analysis (next-generation sequencing) and computational investigation with developmental, microscopy, and genetic analyses (including CRISPR-Cas9 gene editing, tissue-specific RNAi knockdown, etc). We were previously able to assemble the complete genetic framework controlling germline stem cell self-renewal and differentiation in vivo, revealing conserved new aspects of stem cell biology (Teixeira et al, Nature Cell Biology, 2015; Sanchez et al, Cell Stem Cell, 2016). Moving forward, our goal is to build a refined molecular understanding of how protein synthesis control – a new frontier in gene regulation – governs stem cell fate transitions in vivo. Our lab is generously funded by the Wellcome Trust.
Candidates must have experience in a wide range of molecular biology techniques, and prior expertise in next generation sequencing would be an advantage. Experience working with fly genetics is a plus but not required. The successful candidate will be highly motivated, willing to join a young and dynamic research group, have good communication skills, and possess strong problem solving capacities.
To apply online, please follow the link: http://www.jobs.cam.ac.uk/job/15379/
Applications should include a cover letter, Curriculum Vitae, and the contact information of at least two references.
The position start date is flexible. Application deadline: November 17th, 2017.
For an informal discussion about this position, please contact Dr. Felipe Karam Teixeira (fk319@cam.ac.uk).